Akis Linardos
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Layer Ensembles: A Single-Pass Uncertainty Estimation in Deep Learning for Segmentation
Mar 16, 2022



Abstract:Uncertainty estimation in deep learning has become a leading research field in medical image analysis due to the need for safe utilisation of AI algorithms in clinical practice. Most approaches for uncertainty estimation require sampling the network weights multiple times during testing or training multiple networks. This leads to higher training and testing costs in terms of time and computational resources. In this paper, we propose Layer Ensembles, a novel uncertainty estimation method that uses a single network and requires only a single pass to estimate predictive uncertainty of a network. Moreover, we introduce an image-level uncertainty metric, which is more beneficial for segmentation tasks compared to the commonly used pixel-wise metrics such as entropy and variance. We evaluate our approach on 2D and 3D, binary and multi-class medical image segmentation tasks. Our method shows competitive results with state-of-the-art Deep Ensembles, requiring only a single network and a single pass.
A Review of Generative Adversarial Networks in Cancer Imaging: New Applications, New Solutions
Jul 20, 2021



Abstract:Despite technological and medical advances, the detection, interpretation, and treatment of cancer based on imaging data continue to pose significant challenges. These include high inter-observer variability, difficulty of small-sized lesion detection, nodule interpretation and malignancy determination, inter- and intra-tumour heterogeneity, class imbalance, segmentation inaccuracies, and treatment effect uncertainty. The recent advancements in Generative Adversarial Networks (GANs) in computer vision as well as in medical imaging may provide a basis for enhanced capabilities in cancer detection and analysis. In this review, we assess the potential of GANs to address a number of key challenges of cancer imaging, including data scarcity and imbalance, domain and dataset shifts, data access and privacy, data annotation and quantification, as well as cancer detection, tumour profiling and treatment planning. We provide a critical appraisal of the existing literature of GANs applied to cancer imagery, together with suggestions on future research directions to address these challenges. We analyse and discuss 163 papers that apply adversarial training techniques in the context of cancer imaging and elaborate their methodologies, advantages and limitations. With this work, we strive to bridge the gap between the needs of the clinical cancer imaging community and the current and prospective research on GANs in the artificial intelligence community.
Federated Learning for Multi-Center Imaging Diagnostics: A Study in Cardiovascular Disease
Jul 07, 2021
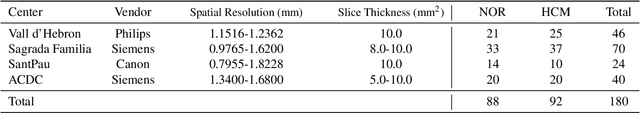

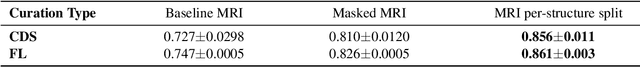
Abstract:Deep learning models can enable accurate and efficient disease diagnosis, but have thus far been hampered by the data scarcity present in the medical world. Automated diagnosis studies have been constrained by underpowered single-center datasets, and although some results have shown promise, their generalizability to other institutions remains questionable as the data heterogeneity between institutions is not taken into account. By allowing models to be trained in a distributed manner that preserves patients' privacy, federated learning promises to alleviate these issues, by enabling diligent multi-center studies. We present the first federated learning study on the modality of cardiovascular magnetic resonance (CMR) and use four centers derived from subsets of the M\&M and ACDC datasets, focusing on the diagnosis of hypertrophic cardiomyopathy (HCM). We adapt a 3D-CNN network pretrained on action recognition and explore two different ways of incorporating shape prior information to the model, and four different data augmentation set-ups, systematically analyzing their impact on the different collaborative learning choices. We show that despite the small size of data (180 subjects derived from four centers), the privacy preserving federated learning achieves promising results that are competitive with traditional centralized learning. We further find that federatively trained models exhibit increased robustness and are more sensitive to domain shift effects.
Calibrated prediction in and out-of-domain for state-of-the-art saliency modeling
May 27, 2021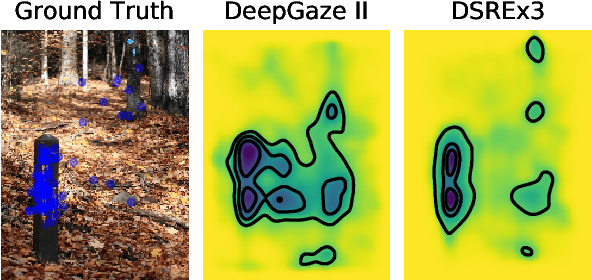
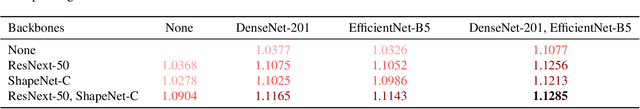
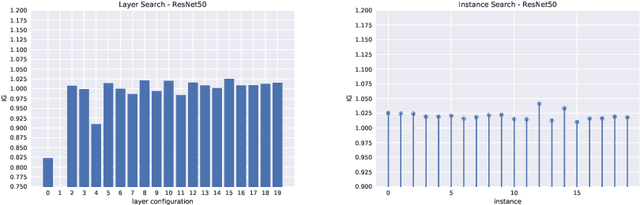
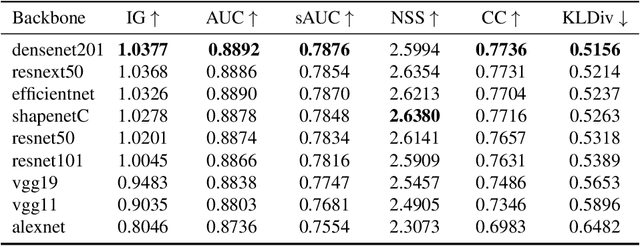
Abstract:Since 2014 transfer learning has become the key driver for the improvement of spatial saliency prediction; however, with stagnant progress in the last 3-5 years. We conduct a large-scale transfer learning study which tests different ImageNet backbones, always using the same read out architecture and learning protocol adopted from DeepGaze II. By replacing the VGG19 backbone of DeepGaze II with ResNet50 features we improve the performance on saliency prediction from 78% to 85%. However, as we continue to test better ImageNet models as backbones (such as EfficientNetB5) we observe no additional improvement on saliency prediction. By analyzing the backbones further, we find that generalization to other datasets differs substantially, with models being consistently overconfident in their fixation predictions. We show that by combining multiple backbones in a principled manner a good confidence calibration on unseen datasets can be achieved. This yields a significant leap in benchmark performance in and out-of-domain with a 15 percent point improvement over DeepGaze II to 93% on MIT1003, marking a new state of the art on the MIT/Tuebingen Saliency Benchmark in all available metrics (AUC: 88.3%, sAUC: 79.4%, CC: 82.4%).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge