Ziyuan Ye
ReLaX: Reasoning with Latent Exploration for Large Reasoning Models
Dec 08, 2025Abstract:Reinforcement Learning with Verifiable Rewards (RLVR) has recently demonstrated remarkable potential in enhancing the reasoning capability of Large Reasoning Models (LRMs). However, RLVR often leads to entropy collapse, resulting in premature policy convergence and performance saturation. While manipulating token-level entropy has proven effective for promoting policy exploration, we argue that the latent dynamics underlying token generation encode a far richer computational structure for steering policy optimization toward a more effective exploration-exploitation tradeoff. To enable tractable analysis and intervention of the latent dynamics of LRMs, we leverage Koopman operator theory to obtain a linearized representation of their hidden-state dynamics. This enables us to introduce Dynamic Spectral Dispersion (DSD), a new metric to quantify the heterogeneity of the model's latent dynamics, serving as a direct indicator of policy exploration. Building upon these foundations, we propose Reasoning with Latent eXploration (ReLaX), a paradigm that explicitly incorporates latent dynamics to regulate exploration and exploitation during policy optimization. Comprehensive experiments across a wide range of multimodal and text-only reasoning benchmarks show that ReLaX significantly mitigates premature convergence and consistently achieves state-of-the-art performance.
Hypergraph Transformer for Semi-Supervised Classification
Dec 18, 2023

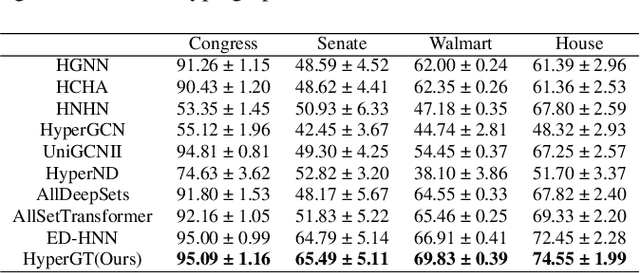
Abstract:Hypergraphs play a pivotal role in the modelling of data featuring higher-order relations involving more than two entities. Hypergraph neural networks emerge as a powerful tool for processing hypergraph-structured data, delivering remarkable performance across various tasks, e.g., hypergraph node classification. However, these models struggle to capture global structural information due to their reliance on local message passing. To address this challenge, we propose a novel hypergraph learning framework, HyperGraph Transformer (HyperGT). HyperGT uses a Transformer-based neural network architecture to effectively consider global correlations among all nodes and hyperedges. To incorporate local structural information, HyperGT has two distinct designs: i) a positional encoding based on the hypergraph incidence matrix, offering valuable insights into node-node and hyperedge-hyperedge interactions; and ii) a hypergraph structure regularization in the loss function, capturing connectivities between nodes and hyperedges. Through these designs, HyperGT achieves comprehensive hypergraph representation learning by effectively incorporating global interactions while preserving local connectivity patterns. Extensive experiments conducted on real-world hypergraph node classification tasks showcase that HyperGT consistently outperforms existing methods, establishing new state-of-the-art benchmarks. Ablation studies affirm the effectiveness of the individual designs of our model.
Explainable fMRI-based Brain Decoding via Spatial Temporal-pyramid Graph Convolutional Network
Oct 08, 2022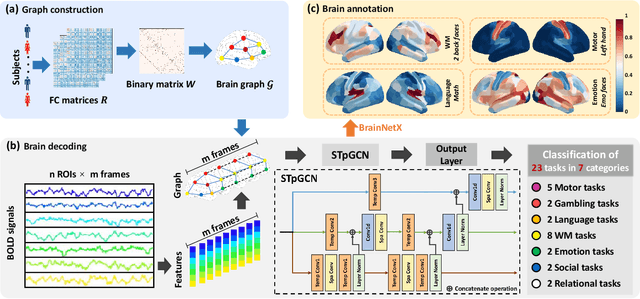
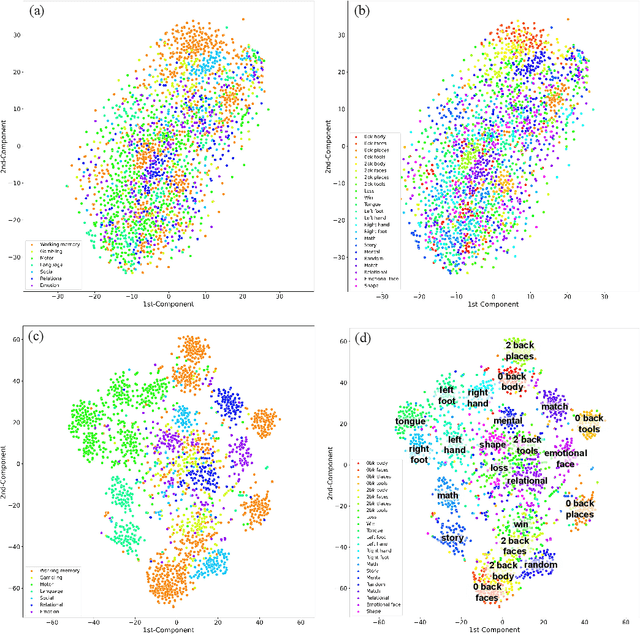
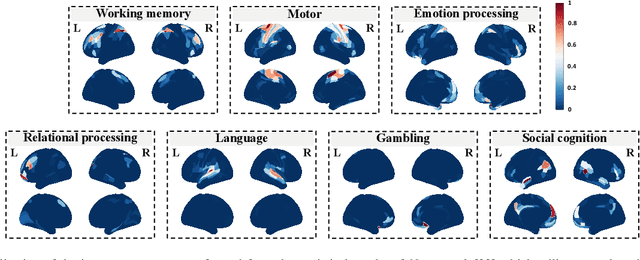
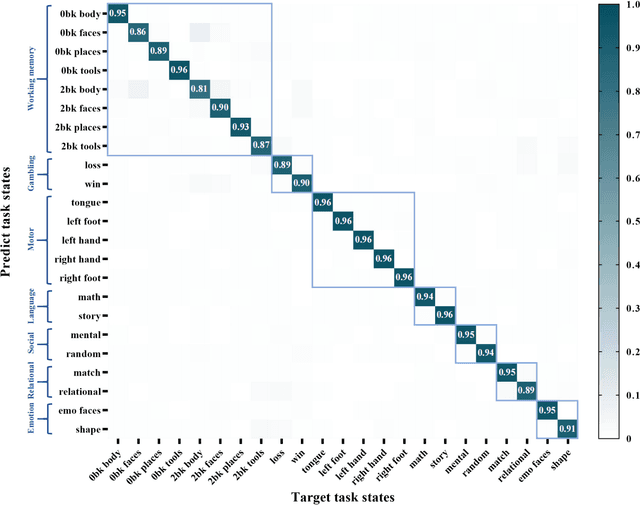
Abstract:Brain decoding, aiming to identify the brain states using neural activity, is important for cognitive neuroscience and neural engineering. However, existing machine learning methods for fMRI-based brain decoding either suffer from low classification performance or poor explainability. Here, we address this issue by proposing a biologically inspired architecture, Spatial Temporal-pyramid Graph Convolutional Network (STpGCN), to capture the spatial-temporal graph representation of functional brain activities. By designing multi-scale spatial-temporal pathways and bottom-up pathways that mimic the information process and temporal integration in the brain, STpGCN is capable of explicitly utilizing the multi-scale temporal dependency of brain activities via graph, thereby achieving high brain decoding performance. Additionally, we propose a sensitivity analysis method called BrainNetX to better explain the decoding results by automatically annotating task-related brain regions from the brain-network standpoint. We conduct extensive experiments on fMRI data under 23 cognitive tasks from Human Connectome Project (HCP) S1200. The results show that STpGCN significantly improves brain decoding performance compared to competing baseline models; BrainNetX successfully annotates task-relevant brain regions. Post hoc analysis based on these regions further validates that the hierarchical structure in STpGCN significantly contributes to the explainability, robustness and generalization of the model. Our methods not only provide insights into information representation in the brain under multiple cognitive tasks but also indicate a bright future for fMRI-based brain decoding.
Immunofluorescence Capillary Imaging Segmentation: Cases Study
Jul 14, 2022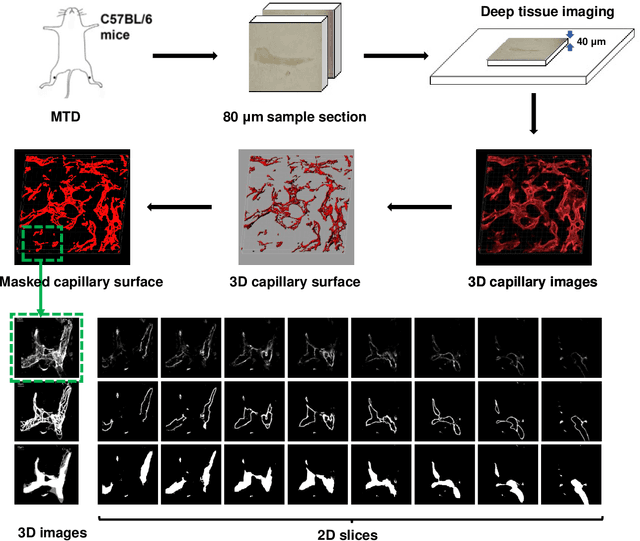
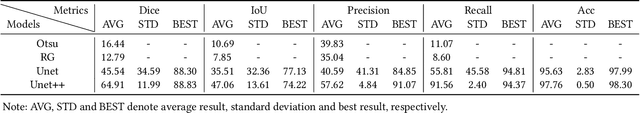
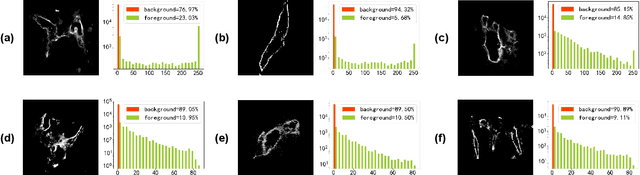
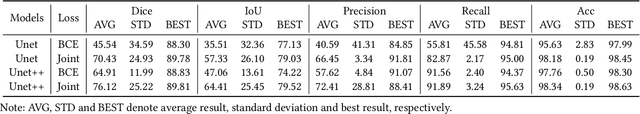
Abstract:Nonunion is one of the challenges faced by orthopedics clinics for the technical difficulties and high costs in photographing interosseous capillaries. Segmenting vessels and filling capillaries are critical in understanding the obstacles encountered in capillary growth. However, existing datasets for blood vessel segmentation mainly focus on the large blood vessels of the body, and the lack of labeled capillary image datasets greatly limits the methodological development and applications of vessel segmentation and capillary filling. Here, we present a benchmark dataset, named IFCIS-155, consisting of 155 2D capillary images with segmentation boundaries and vessel fillings annotated by biomedical experts, and 19 large-scale, high-resolution 3D capillary images. To obtain better images of interosseous capillaries, we leverage state-of-the-art immunofluorescence imaging techniques to highlight the rich vascular morphology of interosseous capillaries. We conduct comprehensive experiments to verify the effectiveness of the dataset and the benchmarking deep learning models (\eg UNet/UNet++ and the modified UNet/UNet++). Our work offers a benchmark dataset for training deep learning models for capillary image segmentation and provides a potential tool for future capillary research. The IFCIS-155 dataset and code are all publicly available at \url{https://github.com/ncclabsustech/IFCIS-55}.
Deep Auto-encoder with Neural Response
Nov 30, 2021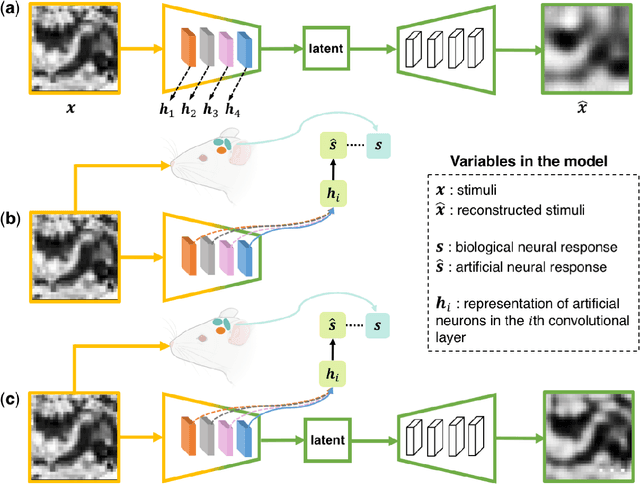
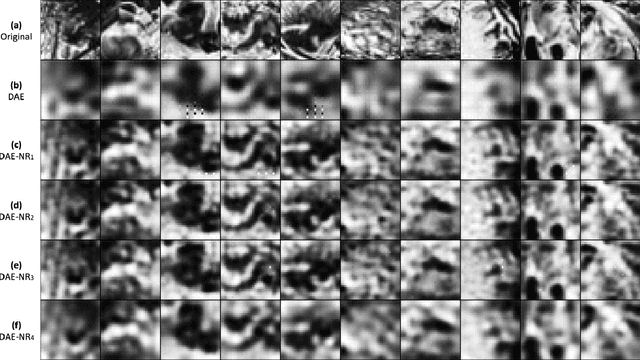
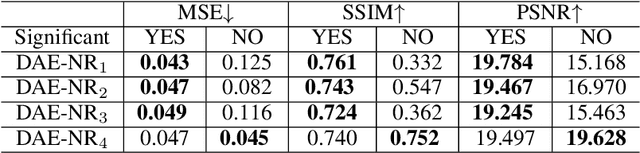
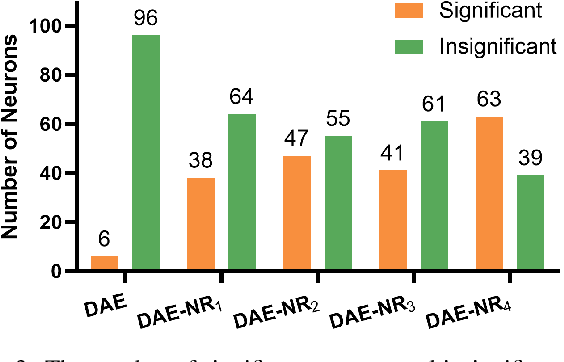
Abstract:Artificial intelligence and neuroscience are deeply interactive. Artificial neural networks (ANNs) have been a versatile tool to study the neural representation in the ventral visual stream, and the knowledge in neuroscience in return inspires ANN models to improve performance in the task. However, how to merge these two directions into a unified model has less studied. Here, we propose a hybrid model, called deep auto-encoder with the neural response (DAE-NR), which incorporates the information from the visual cortex into ANNs to achieve better image reconstruction and higher neural representation similarity between biological and artificial neurons. Specifically, the same visual stimuli (i.e., natural images) are input to both the mice brain and DAE-NR. The DAE-NR jointly learns to map a specific layer of the encoder network to the biological neural responses in the ventral visual stream by a mapping function and to reconstruct the visual input by the decoder. Our experiments demonstrate that if and only if with the joint learning, DAE-NRs can (i) improve the performance of image reconstruction and (ii) increase the representational similarity between biological neurons and artificial neurons. The DAE-NR offers a new perspective on the integration of computer vision and visual neuroscience.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge