Explainable fMRI-based Brain Decoding via Spatial Temporal-pyramid Graph Convolutional Network
Paper and Code
Oct 08, 2022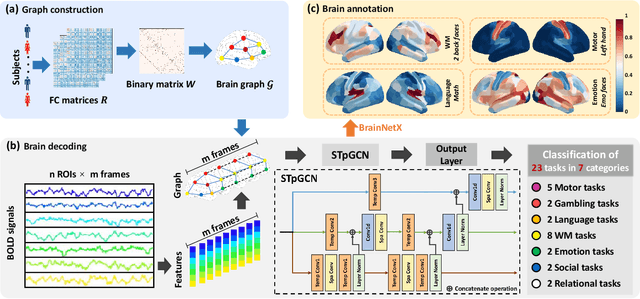
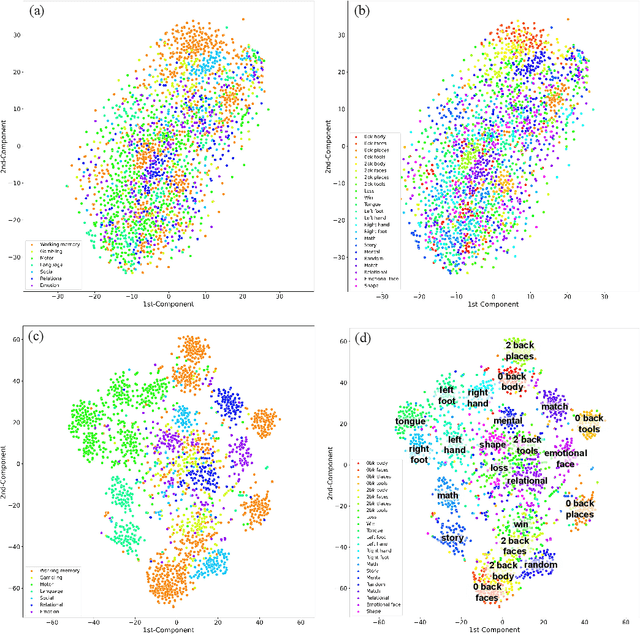
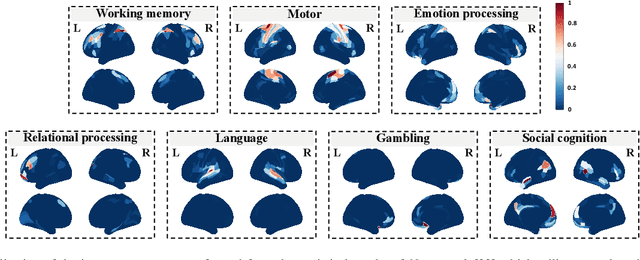
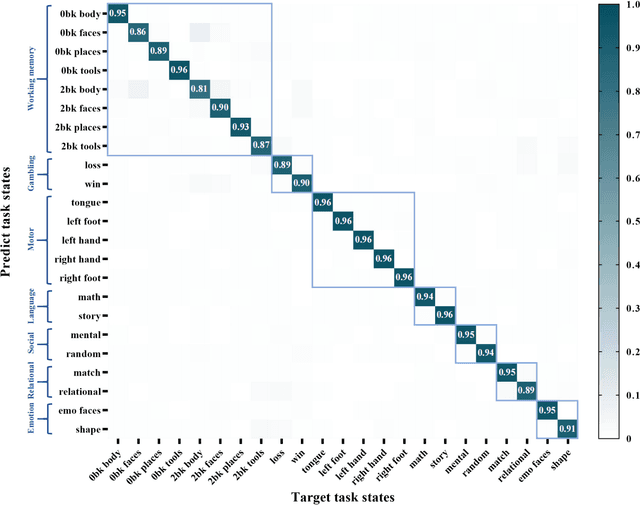
Brain decoding, aiming to identify the brain states using neural activity, is important for cognitive neuroscience and neural engineering. However, existing machine learning methods for fMRI-based brain decoding either suffer from low classification performance or poor explainability. Here, we address this issue by proposing a biologically inspired architecture, Spatial Temporal-pyramid Graph Convolutional Network (STpGCN), to capture the spatial-temporal graph representation of functional brain activities. By designing multi-scale spatial-temporal pathways and bottom-up pathways that mimic the information process and temporal integration in the brain, STpGCN is capable of explicitly utilizing the multi-scale temporal dependency of brain activities via graph, thereby achieving high brain decoding performance. Additionally, we propose a sensitivity analysis method called BrainNetX to better explain the decoding results by automatically annotating task-related brain regions from the brain-network standpoint. We conduct extensive experiments on fMRI data under 23 cognitive tasks from Human Connectome Project (HCP) S1200. The results show that STpGCN significantly improves brain decoding performance compared to competing baseline models; BrainNetX successfully annotates task-relevant brain regions. Post hoc analysis based on these regions further validates that the hierarchical structure in STpGCN significantly contributes to the explainability, robustness and generalization of the model. Our methods not only provide insights into information representation in the brain under multiple cognitive tasks but also indicate a bright future for fMRI-based brain decoding.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge