Ziming Qiu
$μ$-Net: ConvNext-Based U-Nets for Cosmic Muon Tomography
Dec 25, 2023Abstract:Muon scattering tomography utilises muons, typically originating from cosmic rays to image the interiors of dense objects. However, due to the low flux of cosmic ray muons at sea-level and the highly complex interactions that muons display when travelling through matter, existing reconstruction algorithms often suffer from low resolution and high noise. In this work, we develop a novel two-stage deep learning algorithm, $\mu$-Net, consisting of an MLP to predict the muon trajectory and a ConvNeXt-based U-Net to convert the scattering points into voxels. $\mu$-Net achieves a state-of-the-art performance of 17.14 PSNR at the dosage of 1024 muons, outperforming traditional reconstruction algorithms such as the point of closest approach algorithm and maximum likelihood and expectation maximisation algorithm. Furthermore, we find that our method is robust to various corruptions such as inaccuracies in the muon momentum or a limited detector resolution. We also generate and publicly release the first large-scale dataset that maps muon detections to voxels. We hope that our research will spark further investigations into the potential of deep learning to revolutionise this field.
DiffuseExpand: Expanding dataset for 2D medical image segmentation using diffusion models
Apr 26, 2023Abstract:Dataset expansion can effectively alleviate the problem of data scarcity for medical image segmentation, due to privacy concerns and labeling difficulties. However, existing expansion algorithms still face great challenges due to their inability of guaranteeing the diversity of synthesized images with paired segmentation masks. In recent years, Diffusion Probabilistic Models (DPMs) have shown powerful image synthesis performance, even better than Generative Adversarial Networks. Based on this insight, we propose an approach called DiffuseExpand for expanding datasets for 2D medical image segmentation using DPM, which first samples a variety of masks from Gaussian noise to ensure the diversity, and then synthesizes images to ensure the alignment of images and masks. After that, DiffuseExpand chooses high-quality samples to further enhance the effectiveness of data expansion. Our comparison and ablation experiments on COVID-19 and CGMH Pelvis datasets demonstrate the effectiveness of DiffuseExpand. Our code is released at https://anonymous.4open.science/r/DiffuseExpand.
Domain Adaptation of Networks for Camera Pose Estimation: Learning Camera Pose Estimation Without Pose Labels
Nov 29, 2021
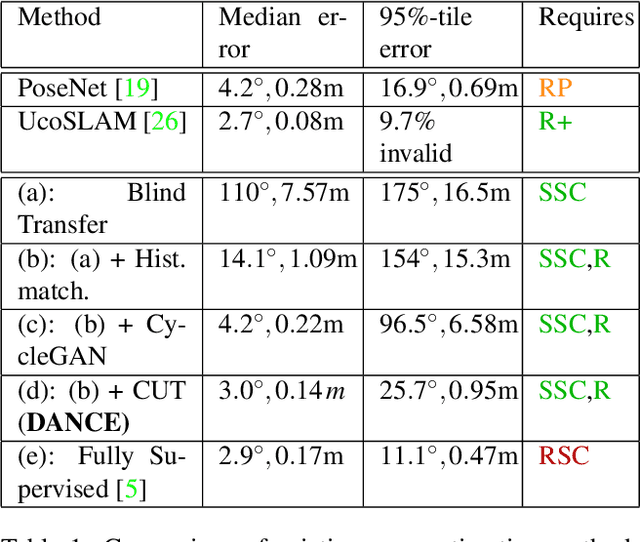


Abstract:One of the key criticisms of deep learning is that large amounts of expensive and difficult-to-acquire training data are required in order to train models with high performance and good generalization capabilities. Focusing on the task of monocular camera pose estimation via scene coordinate regression (SCR), we describe a novel method, Domain Adaptation of Networks for Camera pose Estimation (DANCE), which enables the training of models without access to any labels on the target task. DANCE requires unlabeled images (without known poses, ordering, or scene coordinate labels) and a 3D representation of the space (e.g., a scanned point cloud), both of which can be captured with minimal effort using off-the-shelf commodity hardware. DANCE renders labeled synthetic images from the 3D model, and bridges the inevitable domain gap between synthetic and real images by applying unsupervised image-level domain adaptation techniques (unpaired image-to-image translation). When tested on real images, the SCR model trained with DANCE achieved comparable performance to its fully supervised counterpart (in both cases using PnP-RANSAC for final pose estimation) at a fraction of the cost. Our code and dataset are available at https://github.com/JackLangerman/dance
Deep Mouse: An End-to-end Auto-context Refinement Framework for Brain Ventricle and Body Segmentation in Embryonic Mice Ultrasound Volumes
Oct 30, 2019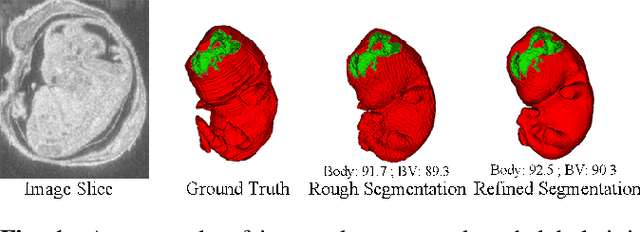
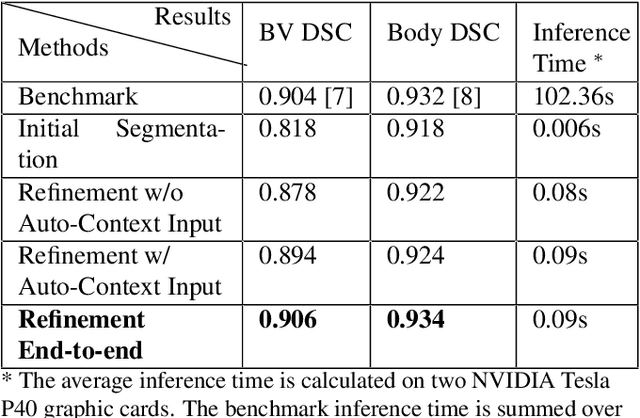
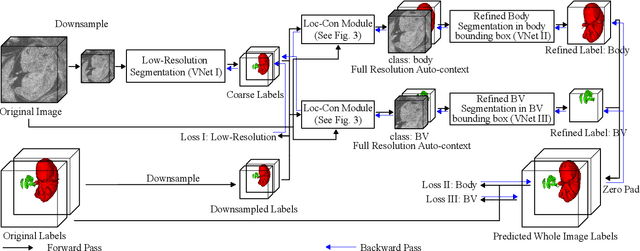
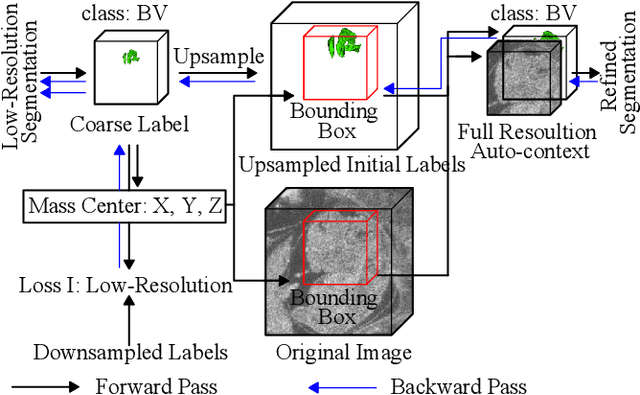
Abstract:High-frequency ultrasound (HFU) is well suited for imaging embryonic mice due to its noninvasive and real-time characteristics. However, manual segmentation of the brain ventricles (BVs) and body requires substantial time and expertise. This work proposes a novel deep learning based end-to-end auto-context refinement framework, consisting of two stages. The first stage produces a low resolution segmentation of the BV and body simultaneously. The resulting probability map for each object (BV or body) is then used to crop a region of interest (ROI) around the target object in both the original image and the probability map to provide context to the refinement segmentation network. Joint training of the two stages provides significant improvement in Dice Similarity Coefficient (DSC) over using only the first stage (0.818 to 0.906 for the BV, and 0.919 to 0.934 for the body). The proposed method significantly reduces the inference time (102.36 to 0.09 s/volume around 1000x faster) while slightly improves the segmentation accuracy over the previous methods using slide-window approaches.
Automatic Mouse Embryo Brain Ventricle & Body Segmentation and Mutant Classification From Ultrasound Data Using Deep Learning
Sep 23, 2019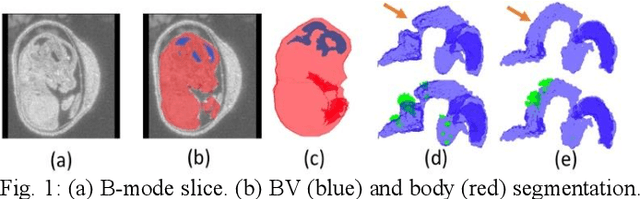
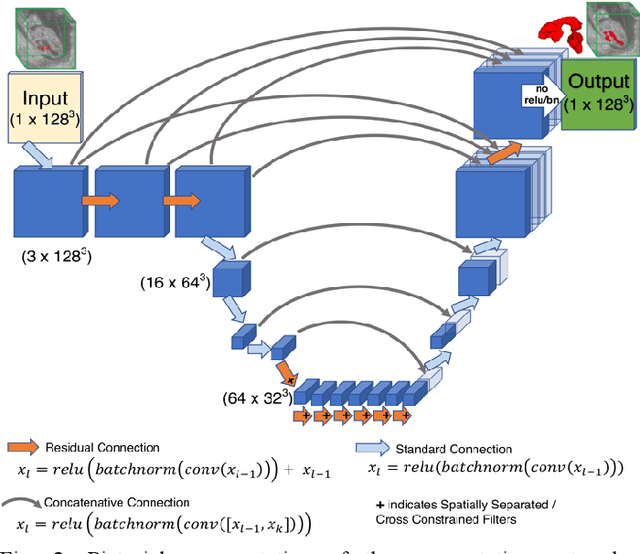
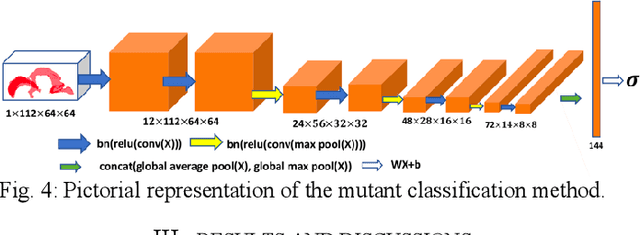
Abstract:High-frequency ultrasound (HFU) is well suited for imaging embryonic mice in vivo because it is non-invasive and real-time. Manual segmentation of the brain ventricles (BVs) and whole body from 3D HFU images is time-consuming and requires specialized training. This paper presents a deep-learning-based segmentation pipeline which automates several time-consuming, repetitive tasks currently performed to study genetic mutations in developing mouse embryos. Namely, the pipeline accurately segments the BV and body regions in 3D HFU images of mouse embryos, despite significant challenges due to position and shape variation of the embryos, as well as imaging artifacts. Based on the BV segmentation, a 3D convolutional neural network (CNN) is further trained to detect embryos with the Engrailed-1 (En1) mutation. The algorithms achieve 0.896 and 0.925 Dice Similarity Coefficient (DSC) for BV and body segmentation, respectively, and 95.8% accuracy on mutant classification. Through gradient based interrogation and visualization of the trained classifier, it is demonstrated that the model focuses on the morphological structures known to be affected by the En1 mutation.
Deep BV: A Fully Automated System for Brain Ventricle Localization and Segmentation in 3D Ultrasound Images of Embryonic Mice
Nov 05, 2018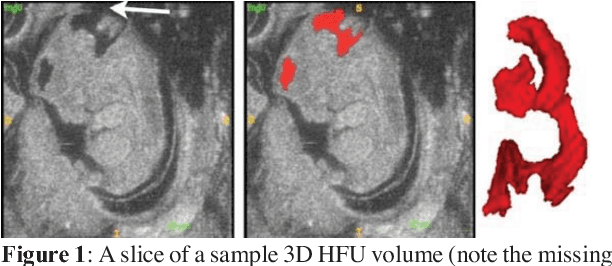
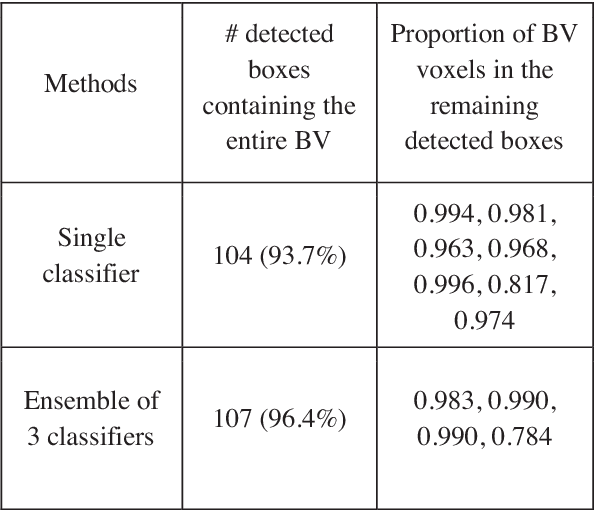
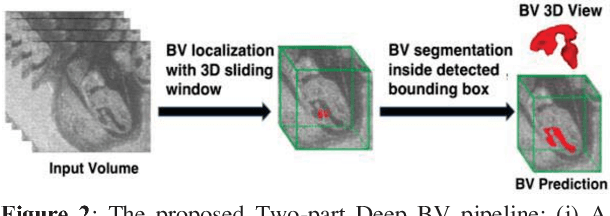
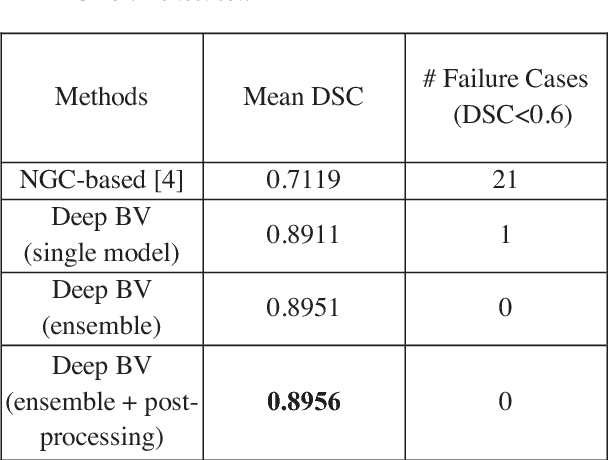
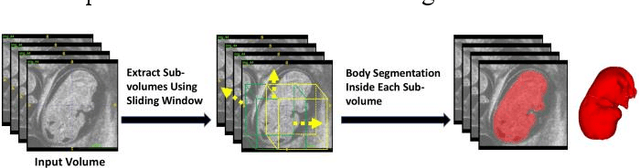
Abstract:Volumetric analysis of brain ventricle (BV) structure is a key tool in the study of central nervous system development in embryonic mice. High-frequency ultrasound (HFU) is the only non-invasive, real-time modality available for rapid volumetric imaging of embryos in utero. However, manual segmentation of the BV from HFU volumes is tedious, time-consuming, and requires specialized expertise. In this paper, we propose a novel deep learning based BV segmentation system for whole-body HFU images of mouse embryos. Our fully automated system consists of two modules: localization and segmentation. It first applies a volumetric convolutional neural network on a 3D sliding window over the entire volume to identify a 3D bounding box containing the entire BV. It then employs a fully convolutional network to segment the detected bounding box into BV and background. The system achieves a Dice Similarity Coefficient (DSC) of 0.8956 for BV segmentation on an unseen 111 HFU volume test set surpassing the previous state-of-the-art method (DSC of 0.7119) by a margin of 25%.
End-to-End Subtitle Detection and Recognition for Videos in East Asian Languages via CNN Ensemble with Near-Human-Level Performance
Nov 18, 2016
Abstract:In this paper, we propose an innovative end-to-end subtitle detection and recognition system for videos in East Asian languages. Our end-to-end system consists of multiple stages. Subtitles are firstly detected by a novel image operator based on the sequence information of consecutive video frames. Then, an ensemble of Convolutional Neural Networks (CNNs) trained on synthetic data is adopted for detecting and recognizing East Asian characters. Finally, a dynamic programming approach leveraging language models is applied to constitute results of the entire body of text lines. The proposed system achieves average end-to-end accuracies of 98.2% and 98.3% on 40 videos in Simplified Chinese and 40 videos in Traditional Chinese respectively, which is a significant outperformance of other existing methods. The near-perfect accuracy of our system dramatically narrows the gap between human cognitive ability and state-of-the-art algorithms used for such a task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge