Jeffrey A. Ketterling
Deep Mouse: An End-to-end Auto-context Refinement Framework for Brain Ventricle and Body Segmentation in Embryonic Mice Ultrasound Volumes
Oct 30, 2019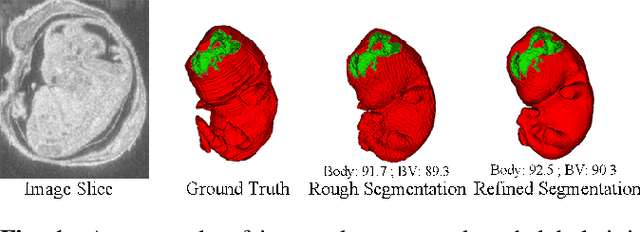
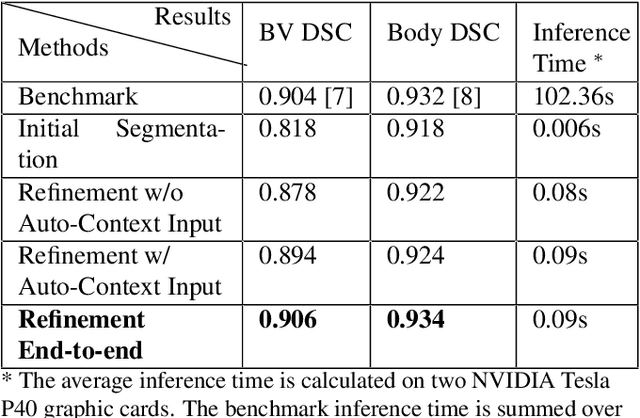
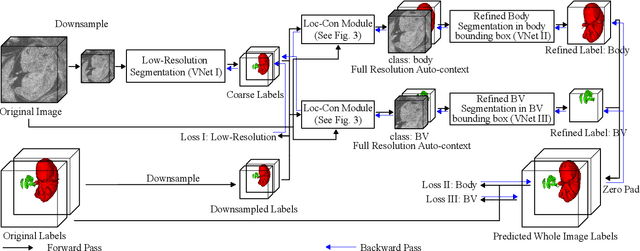
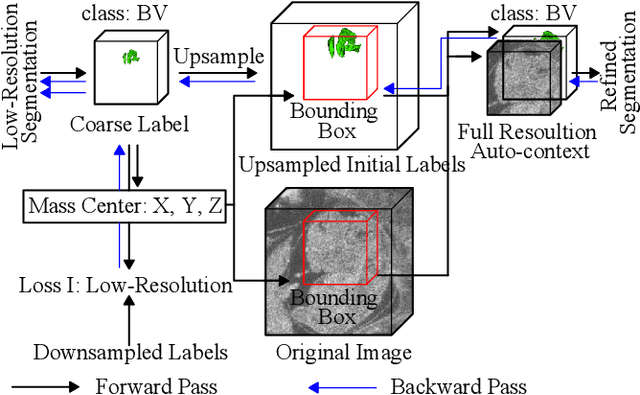
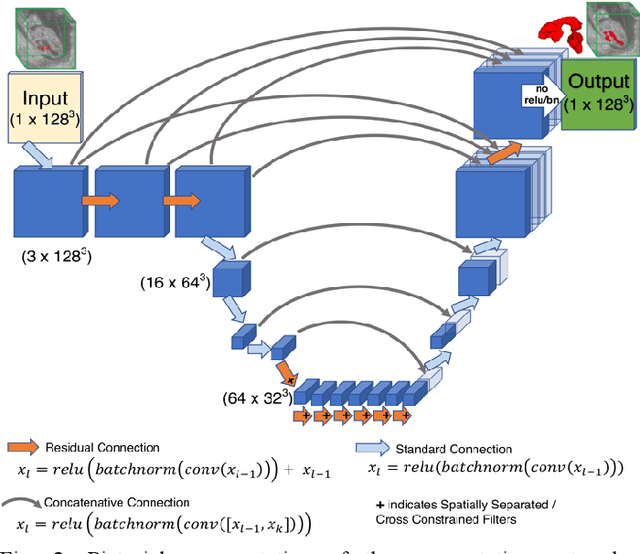
Abstract:High-frequency ultrasound (HFU) is well suited for imaging embryonic mice due to its noninvasive and real-time characteristics. However, manual segmentation of the brain ventricles (BVs) and body requires substantial time and expertise. This work proposes a novel deep learning based end-to-end auto-context refinement framework, consisting of two stages. The first stage produces a low resolution segmentation of the BV and body simultaneously. The resulting probability map for each object (BV or body) is then used to crop a region of interest (ROI) around the target object in both the original image and the probability map to provide context to the refinement segmentation network. Joint training of the two stages provides significant improvement in Dice Similarity Coefficient (DSC) over using only the first stage (0.818 to 0.906 for the BV, and 0.919 to 0.934 for the body). The proposed method significantly reduces the inference time (102.36 to 0.09 s/volume around 1000x faster) while slightly improves the segmentation accuracy over the previous methods using slide-window approaches.
Automatic Mouse Embryo Brain Ventricle & Body Segmentation and Mutant Classification From Ultrasound Data Using Deep Learning
Sep 23, 2019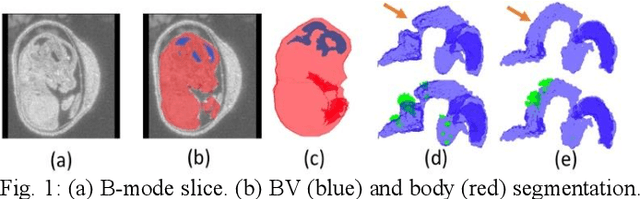
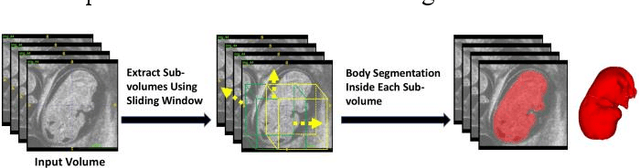
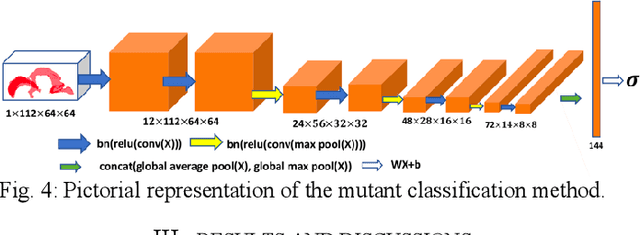
Abstract:High-frequency ultrasound (HFU) is well suited for imaging embryonic mice in vivo because it is non-invasive and real-time. Manual segmentation of the brain ventricles (BVs) and whole body from 3D HFU images is time-consuming and requires specialized training. This paper presents a deep-learning-based segmentation pipeline which automates several time-consuming, repetitive tasks currently performed to study genetic mutations in developing mouse embryos. Namely, the pipeline accurately segments the BV and body regions in 3D HFU images of mouse embryos, despite significant challenges due to position and shape variation of the embryos, as well as imaging artifacts. Based on the BV segmentation, a 3D convolutional neural network (CNN) is further trained to detect embryos with the Engrailed-1 (En1) mutation. The algorithms achieve 0.896 and 0.925 Dice Similarity Coefficient (DSC) for BV and body segmentation, respectively, and 95.8% accuracy on mutant classification. Through gradient based interrogation and visualization of the trained classifier, it is demonstrated that the model focuses on the morphological structures known to be affected by the En1 mutation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge