Zhigang Song
Fusion of Heterogeneous Pathology Foundation Models for Whole Slide Image Analysis
Oct 31, 2025Abstract:Whole slide image (WSI) analysis has emerged as an increasingly essential technique in computational pathology. Recent advances in the pathological foundation models (FMs) have demonstrated significant advantages in deriving meaningful patch-level or slide-level feature representations from WSIs. However, current pathological FMs have exhibited substantial heterogeneity caused by diverse private training datasets and different network architectures. This heterogeneity introduces performance variability when we utilize the extracted features from different FMs in the downstream tasks. To fully explore the advantage of multiple FMs effectively, in this work, we propose a novel framework for the fusion of heterogeneous pathological FMs, called FuseCPath, yielding a model with a superior ensemble performance. The main contributions of our framework can be summarized as follows: (i) To guarantee the representativeness of the training patches, we propose a multi-view clustering-based method to filter out the discriminative patches via multiple FMs' embeddings. (ii) To effectively fuse the heterogeneous patch-level FMs, we devise a cluster-level re-embedding strategy to online capture patch-level local features. (iii) To effectively fuse the heterogeneous slide-level FMs, we devise a collaborative distillation strategy to explore the connections between slide-level FMs. Extensive experiments conducted on lung cancer, bladder cancer, and colorectal cancer datasets from The Cancer Genome Atlas (TCGA) have demonstrated that the proposed FuseCPath achieves state-of-the-art performance across multiple tasks on these public datasets.
CAMEL2: Enhancing weakly supervised learning for histopathology images by incorporating the significance ratio
Oct 09, 2023Abstract:Histopathology image analysis plays a crucial role in cancer diagnosis. However, training a clinically applicable segmentation algorithm requires pathologists to engage in labour-intensive labelling. In contrast, weakly supervised learning methods, which only require coarse-grained labels at the image level, can significantly reduce the labeling efforts. Unfortunately, while these methods perform reasonably well in slide-level prediction, their ability to locate cancerous regions, which is essential for many clinical applications, remains unsatisfactory. Previously, we proposed CAMEL, which achieves comparable results to those of fully supervised baselines in pixel-level segmentation. However, CAMEL requires 1,280x1,280 image-level binary annotations for positive WSIs. Here, we present CAMEL2, by introducing a threshold of the cancerous ratio for positive bags, it allows us to better utilize the information, consequently enabling us to scale up the image-level setting from 1,280x1,280 to 5,120x5,120 while maintaining the accuracy. Our results with various datasets, demonstrate that CAMEL2, with the help of 5,120x5,120 image-level binary annotations, which are easy to annotate, achieves comparable performance to that of a fully supervised baseline in both instance- and slide-level classifications.
MACCIF-TDNN: Multi aspect aggregation of channel and context interdependence features in TDNN-based speaker verification
Jul 07, 2021



Abstract:Most of the recent state-of-the-art results for speaker verification are achieved by X-vector and its subsequent variants. In this paper, we propose a new network architecture which aggregates the channel and context interdependence features from multi aspect based on Time Delay Neural Network (TDNN). Firstly, we use the SE-Res2Blocks as in ECAPA-TDNN to explicitly model the channel interdependence to realize adaptive calibration of channel features, and process local context features in a multi-scale way at a more granular level compared with conventional TDNN-based methods. Secondly, we explore to use the encoder structure of Transformer to model the global context interdependence features at an utterance level which can capture better long term temporal characteristics. Before the pooling layer, we aggregate the outputs of SE-Res2Blocks and Transformer encoder to leverage the complementary channel and context interdependence features learned by themself respectively. Finally, instead of performing a single attentive statistics pooling, we also find it beneficial to extend the pooling method in a multi-head way which can discriminate features from multiple aspect. The proposed MACCIF-TDNN architecture can outperform most of the state-of-the-art TDNN-based systems on VoxCeleb1 test sets.
CAMEL: A Weakly Supervised Learning Framework for Histopathology Image Segmentation
Aug 28, 2019
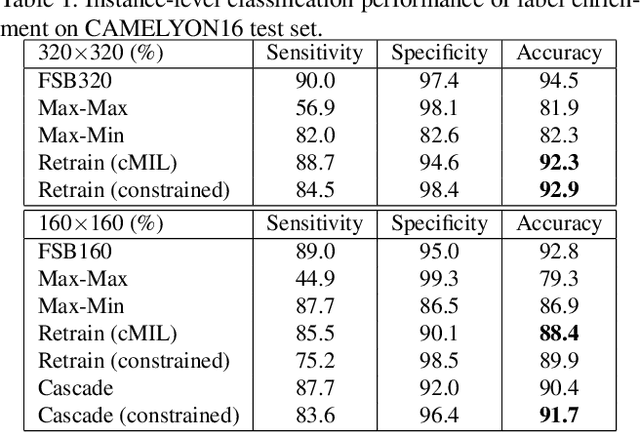


Abstract:Histopathology image analysis plays a critical role in cancer diagnosis and treatment. To automatically segment the cancerous regions, fully supervised segmentation algorithms require labor-intensive and time-consuming labeling at the pixel level. In this research, we propose CAMEL, a weakly supervised learning framework for histopathology image segmentation using only image-level labels. Using multiple instance learning (MIL)-based label enrichment, CAMEL splits the image into latticed instances and automatically generates instance-level labels. After label enrichment, the instance-level labels are further assigned to the corresponding pixels, producing the approximate pixel-level labels and making fully supervised training of segmentation models possible. CAMEL achieves comparable performance with the fully supervised approaches in both instance-level classification and pixel-level segmentation on CAMELYON16 and a colorectal adenoma dataset. Moreover, the generality of the automatic labeling methodology may benefit future weakly supervised learning studies for histopathology image analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge