Shuhao Wang
CAMEL2: Enhancing weakly supervised learning for histopathology images by incorporating the significance ratio
Oct 09, 2023Abstract:Histopathology image analysis plays a crucial role in cancer diagnosis. However, training a clinically applicable segmentation algorithm requires pathologists to engage in labour-intensive labelling. In contrast, weakly supervised learning methods, which only require coarse-grained labels at the image level, can significantly reduce the labeling efforts. Unfortunately, while these methods perform reasonably well in slide-level prediction, their ability to locate cancerous regions, which is essential for many clinical applications, remains unsatisfactory. Previously, we proposed CAMEL, which achieves comparable results to those of fully supervised baselines in pixel-level segmentation. However, CAMEL requires 1,280x1,280 image-level binary annotations for positive WSIs. Here, we present CAMEL2, by introducing a threshold of the cancerous ratio for positive bags, it allows us to better utilize the information, consequently enabling us to scale up the image-level setting from 1,280x1,280 to 5,120x5,120 while maintaining the accuracy. Our results with various datasets, demonstrate that CAMEL2, with the help of 5,120x5,120 image-level binary annotations, which are easy to annotate, achieves comparable performance to that of a fully supervised baseline in both instance- and slide-level classifications.
Automated Scoring System of HER2 in Pathological Images under the Microscope
Oct 21, 2021
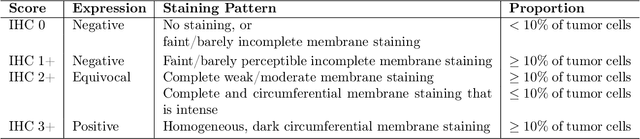
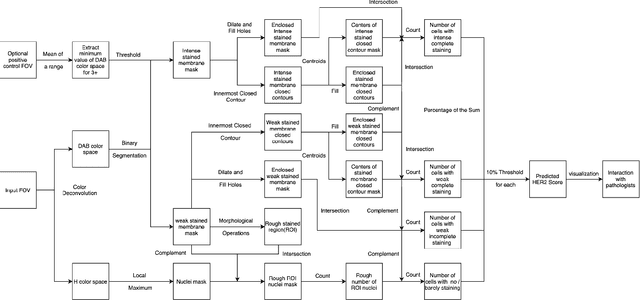

Abstract:Breast cancer is the most common cancer among women worldwide. The human epidermal growth factor receptor 2(HER2) with immunohistochemical(IHC) is widely used for pathological evaluation to provide the appropriate therapy for patients with breast cancer. However, the deficiency of pathologists is extremely significant in the current society, and visual diagnosis of the HER2 overexpression is subjective and susceptible to inter-observer variation. Recently, with the rapid development of artificial intelligence(AI) in disease diagnosis, several automated HER2 scoring methods using traditional computer vision or machine learning methods indicate the improvement of the HER2 diagnostic accuracy, but the unreasonable interpretation in pathology, as well as the expensive and ethical issues for annotation, make these methods still have a long way to deploy in hospitals to ease pathologists' burden in real. In this paper, we propose a HER2 automated scoring system that strictly follows the HER2 scoring guidelines simulating the real workflow of HER2 scores diagnosis by pathologists. Unlike the previous work, our method takes the positive control of HER2 into account to make sure the assay performance for each slide, eliminating work for repeated comparison and checking for the current field of view(FOV) and positive control FOV, especially for the borderline cases. Besides, for each selected FOV under the microscope, our system provides real-time HER2 scores analysis and visualizations of the membrane staining intensity and completeness corresponding with the cell classification. Our rigorous workflow along with the flexible interactive adjustion in demand substantially assists pathologists to finish the HER2 diagnosis faster and improves the robustness and accuracy. The proposed system will be embedded in our Thorough Eye platform for deployment in hospitals.
CAMEL: A Weakly Supervised Learning Framework for Histopathology Image Segmentation
Aug 28, 2019
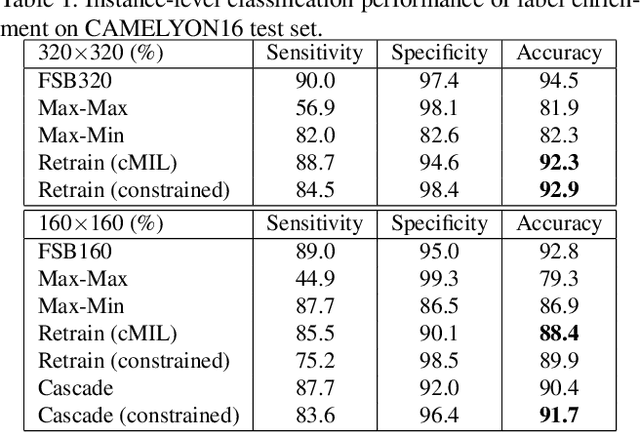


Abstract:Histopathology image analysis plays a critical role in cancer diagnosis and treatment. To automatically segment the cancerous regions, fully supervised segmentation algorithms require labor-intensive and time-consuming labeling at the pixel level. In this research, we propose CAMEL, a weakly supervised learning framework for histopathology image segmentation using only image-level labels. Using multiple instance learning (MIL)-based label enrichment, CAMEL splits the image into latticed instances and automatically generates instance-level labels. After label enrichment, the instance-level labels are further assigned to the corresponding pixels, producing the approximate pixel-level labels and making fully supervised training of segmentation models possible. CAMEL achieves comparable performance with the fully supervised approaches in both instance-level classification and pixel-level segmentation on CAMELYON16 and a colorectal adenoma dataset. Moreover, the generality of the automatic labeling methodology may benefit future weakly supervised learning studies for histopathology image analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge