CAMEL: A Weakly Supervised Learning Framework for Histopathology Image Segmentation
Paper and Code
Aug 28, 2019
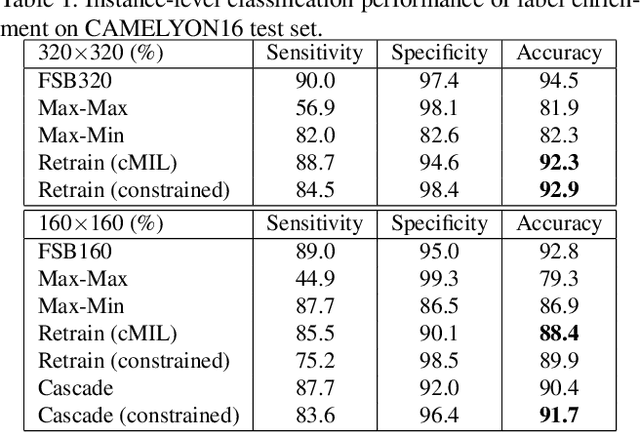


Histopathology image analysis plays a critical role in cancer diagnosis and treatment. To automatically segment the cancerous regions, fully supervised segmentation algorithms require labor-intensive and time-consuming labeling at the pixel level. In this research, we propose CAMEL, a weakly supervised learning framework for histopathology image segmentation using only image-level labels. Using multiple instance learning (MIL)-based label enrichment, CAMEL splits the image into latticed instances and automatically generates instance-level labels. After label enrichment, the instance-level labels are further assigned to the corresponding pixels, producing the approximate pixel-level labels and making fully supervised training of segmentation models possible. CAMEL achieves comparable performance with the fully supervised approaches in both instance-level classification and pixel-level segmentation on CAMELYON16 and a colorectal adenoma dataset. Moreover, the generality of the automatic labeling methodology may benefit future weakly supervised learning studies for histopathology image analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge