Zhenglin Chen
StructCore: Structure-Aware Image-Level Scoring for Training-Free Unsupervised Anomaly Detection
Feb 19, 2026Abstract:Max pooling is the de facto standard for converting anomaly score maps into image-level decisions in memory-bank-based unsupervised anomaly detection (UAD). However, because it relies on a single extreme response, it discards most information about how anomaly evidence is distributed and structured across the image, often causing normal and anomalous scores to overlap. We propose StructCore, a training-free, structure-aware image-level scoring method that goes beyond max pooling. Given an anomaly score map, StructCore computes a low-dimensional structural descriptor phi(S) that captures distributional and spatial characteristics, and refines image-level scoring via a diagonal Mahalanobis calibration estimated from train-good samples, without modifying pixel-level localization. StructCore achieves image-level AUROC scores of 99.6% on MVTec AD and 98.4% on VisA, demonstrating robust image-level anomaly detection by exploiting structural signatures missed by max pooling.
Mapping the maturation of TCM as an adjuvant to radiotherapy
Jan 17, 2026Abstract:The integration of complementary medicine into oncology represents a paradigm shift that has seen to increasing adoption of Traditional Chinese Medicine (TCM) as an adjuvant to radiotherapy. About twenty-five years since the formal institutionalization of integrated oncology, it is opportune to synthesize the trajectory of evidence for TCM as an adjuvant to radiotherapy. Here we conduct a large-scale analysis of 69,745 publications (2000 - 2025), emerging a cyclical evolution defined by coordinated expansion and contraction in publication output, international collaboration, and funding commitments that mirrors a define-ideate-test pattern. Using a theme modeling workflow designed to determine a stable thematic structure of the field, we identify five dominant thematic axes - cancer types, supportive care, clinical endpoints, mechanisms, and methodology - that signal a focus on patient well-being, scientific rigor and mechanistic exploration. Cross-theme integration of TCM is patient-centered and systems-oriented. Together with the emergent cycles of evolution, the thematic structure demonstrates progressive specialization and potential defragmentation of the field or saturation of existing research agenda. The analysis points to a field that has matured its current research agenda and is likely at the cusp of something new. Additionally, the field exhibits positive reporting of findings that is homogeneous across publication types, thematic areas, and the cycles of evolution suggesting a system-wide positive reporting bias agnostic to structural drivers.
GCR: Geometry-Consistent Routing for Task-Agnostic Continual Anomaly Detection
Jan 08, 2026Abstract:Feature-based anomaly detection is widely adopted in industrial inspection due to the strong representational power of large pre-trained vision encoders. While most existing methods focus on improving within-category anomaly scoring, practical deployments increasingly require task-agnostic operation under continual category expansion, where the category identity is unknown at test time. In this setting, overall performance is often dominated by expert selection, namely routing an input to an appropriate normality model before any head-specific scoring is applied. However, routing rules that compare head-specific anomaly scores across independently constructed heads are unreliable in practice, as score distributions can differ substantially across categories in scale and tail behavior. We propose GCR, a lightweight mixture-of-experts framework for stabilizing task-agnostic continual anomaly detection through geometry-consistent routing. GCR routes each test image directly in a shared frozen patch-embedding space by minimizing an accumulated nearest-prototype distance to category-specific prototype banks, and then computes anomaly maps only within the routed expert using a standard prototype-based scoring rule. By separating cross-head decision making from within-head anomaly scoring, GCR avoids cross-head score comparability issues without requiring end-to-end representation learning. Experiments on MVTec AD and VisA show that geometry-consistent routing substantially improves routing stability and mitigates continual performance collapse, achieving near-zero forgetting while maintaining competitive detection and localization performance. These results indicate that many failures previously attributed to representation forgetting can instead be explained by decision-rule instability in cross-head routing. Code is available at https://github.com/jw-chae/GCR
HCMA-UNet: A Hybrid CNN-Mamba UNet with Inter-Slice Self-Attention for Efficient Breast Cancer Segmentation
Jan 01, 2025



Abstract:Breast cancer lesion segmentation in DCE-MRI remains challenging due to heterogeneous tumor morphology and indistinct boundaries. To address these challenges, this study proposes a novel hybrid segmentation network, HCMA-UNet, for lesion segmentation of breast cancer. Our network consists of a lightweight CNN backbone and a Multi-view Inter-Slice Self-Attention Mamba (MISM) module. The MISM module integrates Visual State Space Block (VSSB) and Inter-Slice Self-Attention (ISSA) mechanism, effectively reducing parameters through Asymmetric Split Channel (ASC) strategy to achieve efficient tri-directional feature extraction. Our lightweight model achieves superior performance with 2.87M parameters and 126.44 GFLOPs. A Feature-guided Region-aware loss function (FRLoss) is proposed to enhance segmentation accuracy. Extensive experiments on one private and two public DCE-MRI breast cancer datasets demonstrate that our approach achieves state-of-the-art performance while maintaining computational efficiency. FRLoss also exhibits good cross-architecture generalization capabilities. The source code and dataset is available on this link.
Benchmarking the Cell Image Segmentation Models Robustness under the Microscope Optical Aberrations
Apr 12, 2024Abstract:Cell segmentation is essential in biomedical research for analyzing cellular morphology and behavior. Deep learning methods, particularly convolutional neural networks (CNNs), have revolutionized cell segmentation by extracting intricate features from images. However, the robustness of these methods under microscope optical aberrations remains a critical challenge. This study comprehensively evaluates the performance of cell instance segmentation models under simulated aberration conditions using the DynamicNuclearNet (DNN) and LIVECell datasets. Aberrations, including Astigmatism, Coma, Spherical, and Trefoil, were simulated using Zernike polynomial equations. Various segmentation models, such as Mask R-CNN with different network heads (FPN, C3) and backbones (ResNet, VGG19, SwinS), were trained and tested under aberrated conditions. Results indicate that FPN combined with SwinS demonstrates superior robustness in handling simple cell images affected by minor aberrations. Conversely, Cellpose2.0 proves effective for complex cell images under similar conditions. Our findings provide insights into selecting appropriate segmentation models based on cell morphology and aberration severity, enhancing the reliability of cell segmentation in biomedical applications. Further research is warranted to validate these methods with diverse aberration types and emerging segmentation models. Overall, this research aims to guide researchers in effectively utilizing cell segmentation models in the presence of minor optical aberrations.
Pneumonia App: a mobile application for efficient pediatric pneumonia diagnosis using explainable convolutional neural networks (CNN)
Mar 31, 2024



Abstract:Mycoplasma pneumoniae pneumonia (MPP) poses significant diagnostic challenges in pediatric healthcare, especially in regions like China where it's prevalent. We introduce PneumoniaAPP, a mobile application leveraging deep learning techniques for rapid MPP detection. Our approach capitalizes on convolutional neural networks (CNNs) trained on a comprehensive dataset comprising 3345 chest X-ray (CXR) images, which includes 833 CXR images revealing MPP and additionally augmented with samples from a public dataset. The CNN model achieved an accuracy of 88.20% and an AUROC of 0.9218 across all classes, with a specific accuracy of 97.64% for the mycoplasma class, as demonstrated on the testing dataset. Furthermore, we integrated explainability techniques into PneumoniaAPP to aid respiratory physicians in lung opacity localization. Our contribution extends beyond existing research by targeting pediatric MPP, emphasizing the age group of 0-12 years, and prioritizing deployment on mobile devices. This work signifies a significant advancement in pediatric pneumonia diagnosis, offering a reliable and accessible tool to alleviate diagnostic burdens in healthcare settings.
Harnessing Intra-group Variations Via a Population-Level Context for Pathology Detection
Mar 04, 2024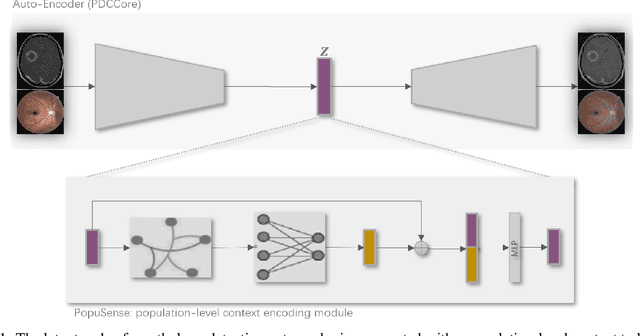
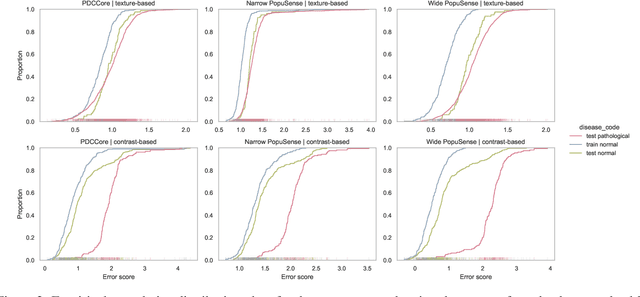
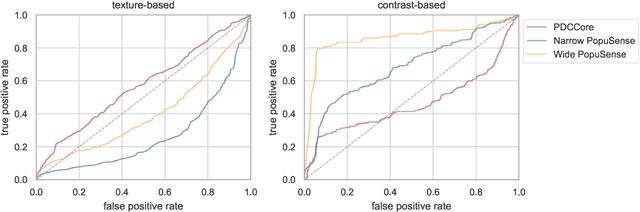
Abstract:Realizing sufficient separability between the distributions of healthy and pathological samples is a critical obstacle for pathology detection convolutional models. Moreover, these models exhibit a bias for contrast-based images, with diminished performance on texture-based medical images. This study introduces the notion of a population-level context for pathology detection and employs a graph theoretic approach to model and incorporate it into the latent code of an autoencoder via a refinement module we term PopuSense. PopuSense seeks to capture additional intra-group variations inherent in biomedical data that a local or global context of the convolutional model might miss or smooth out. Experiments on contrast-based and texture-based images, with minimal adaptation, encounter the existing preference for intensity-based input. Nevertheless, PopuSense demonstrates improved separability in contrast-based images, presenting an additional avenue for refining representations learned by a model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge