Zelin Zhang
EGFormer: Towards Efficient and Generalizable Multimodal Semantic Segmentation
May 20, 2025Abstract:Recent efforts have explored multimodal semantic segmentation using various backbone architectures. However, while most methods aim to improve accuracy, their computational efficiency remains underexplored. To address this, we propose EGFormer, an efficient multimodal semantic segmentation framework that flexibly integrates an arbitrary number of modalities while significantly reducing model parameters and inference time without sacrificing performance. Our framework introduces two novel modules. First, the Any-modal Scoring Module (ASM) assigns importance scores to each modality independently, enabling dynamic ranking based on their feature maps. Second, the Modal Dropping Module (MDM) filters out less informative modalities at each stage, selectively preserving and aggregating only the most valuable features. This design allows the model to leverage useful information from all available modalities while discarding redundancy, thus ensuring high segmentation quality. In addition to efficiency, we evaluate EGFormer on a synthetic-to-real transfer task to demonstrate its generalizability. Extensive experiments show that EGFormer achieves competitive performance with up to 88 percent reduction in parameters and 50 percent fewer GFLOPs. Under unsupervised domain adaptation settings, it further achieves state-of-the-art transfer performance compared to existing methods.
BCDDM: Branch-Corrected Denoising Diffusion Model for Black Hole Image Generation
Feb 12, 2025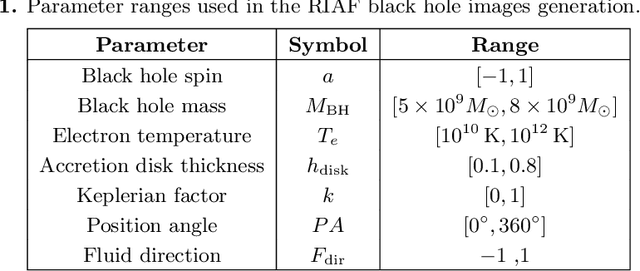
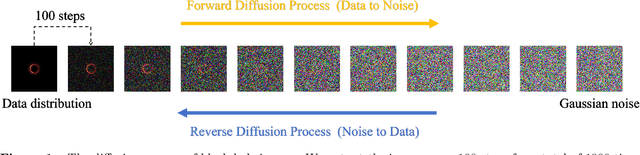
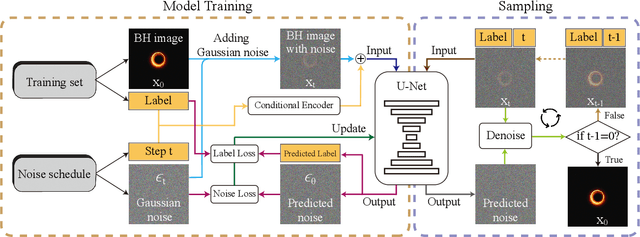
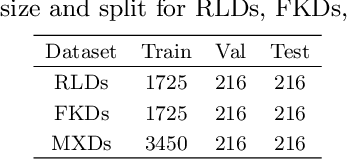
Abstract:The properties of black holes and accretion flows can be inferred by fitting Event Horizon Telescope (EHT) data to simulated images generated through general relativistic ray tracing (GRRT). However, due to the computationally intensive nature of GRRT, the efficiency of generating specific radiation flux images needs to be improved. This paper introduces the Branch Correction Denoising Diffusion Model (BCDDM), which uses a branch correction mechanism and a weighted mixed loss function to improve the accuracy of generated black hole images based on seven physical parameters of the radiatively inefficient accretion flow (RIAF) model. Our experiments show a strong correlation between the generated images and their physical parameters. By enhancing the GRRT dataset with BCDDM-generated images and using ResNet50 for parameter regression, we achieve significant improvements in parameter prediction performance. This approach reduces computational costs and provides a faster, more efficient method for dataset expansion, parameter estimation, and model fitting.
Fetal Brain Tissue Annotation and Segmentation Challenge Results
Apr 20, 2022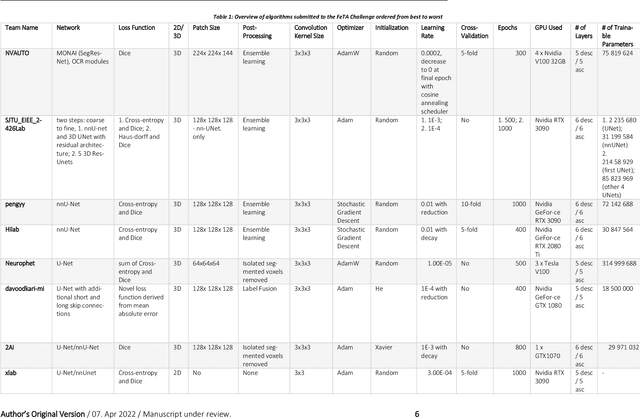
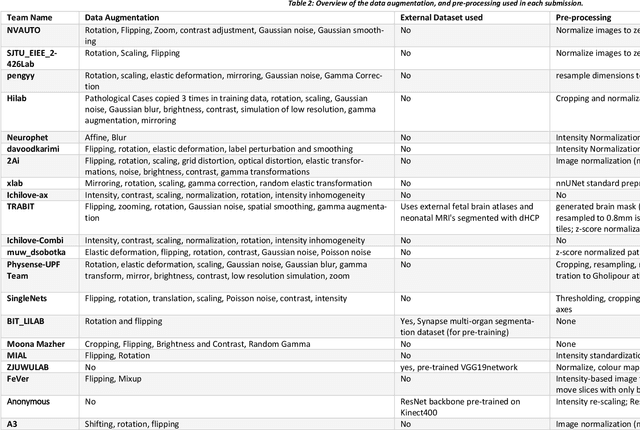
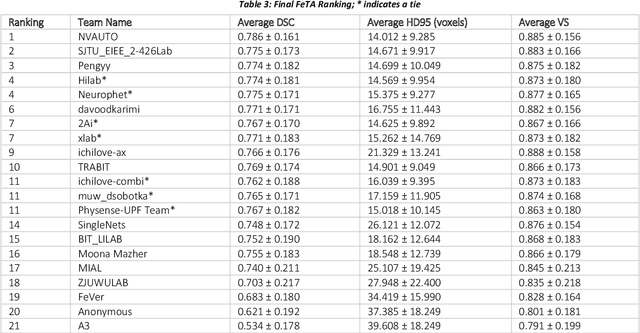
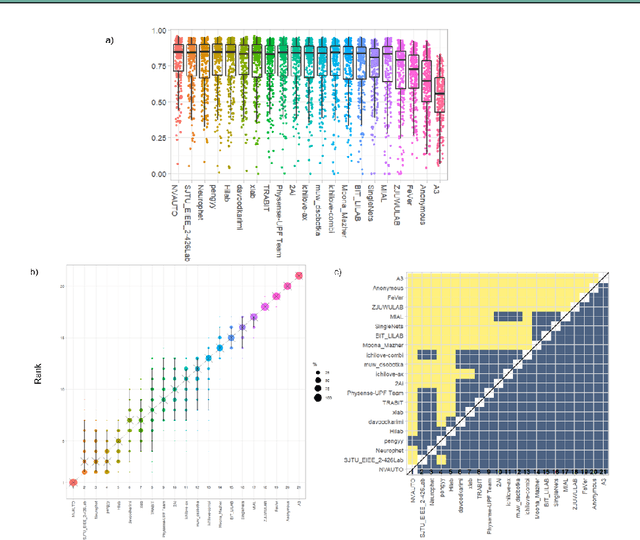
Abstract:In-utero fetal MRI is emerging as an important tool in the diagnosis and analysis of the developing human brain. Automatic segmentation of the developing fetal brain is a vital step in the quantitative analysis of prenatal neurodevelopment both in the research and clinical context. However, manual segmentation of cerebral structures is time-consuming and prone to error and inter-observer variability. Therefore, we organized the Fetal Tissue Annotation (FeTA) Challenge in 2021 in order to encourage the development of automatic segmentation algorithms on an international level. The challenge utilized FeTA Dataset, an open dataset of fetal brain MRI reconstructions segmented into seven different tissues (external cerebrospinal fluid, grey matter, white matter, ventricles, cerebellum, brainstem, deep grey matter). 20 international teams participated in this challenge, submitting a total of 21 algorithms for evaluation. In this paper, we provide a detailed analysis of the results from both a technical and clinical perspective. All participants relied on deep learning methods, mainly U-Nets, with some variability present in the network architecture, optimization, and image pre- and post-processing. The majority of teams used existing medical imaging deep learning frameworks. The main differences between the submissions were the fine tuning done during training, and the specific pre- and post-processing steps performed. The challenge results showed that almost all submissions performed similarly. Four of the top five teams used ensemble learning methods. However, one team's algorithm performed significantly superior to the other submissions, and consisted of an asymmetrical U-Net network architecture. This paper provides a first of its kind benchmark for future automatic multi-tissue segmentation algorithms for the developing human brain in utero.
Image Reconstruction from Events. Why learn it?
Dec 12, 2021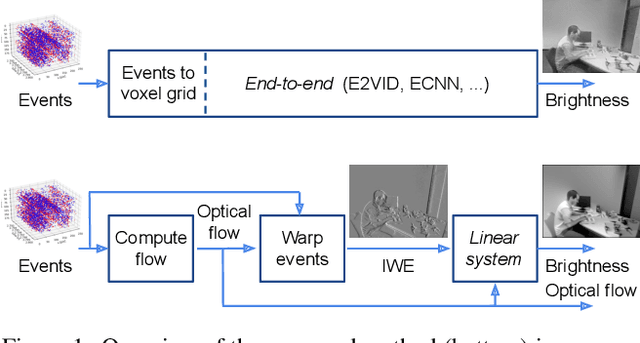
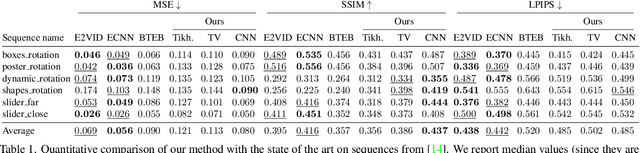
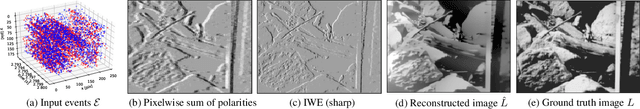
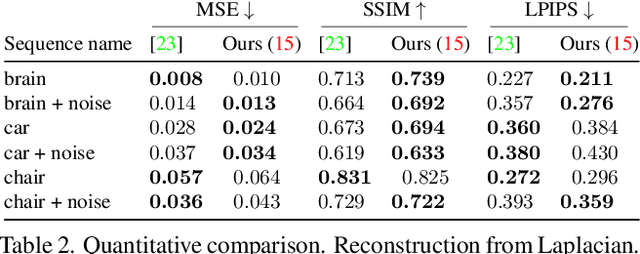
Abstract:Traditional cameras measure image intensity. Event cameras, by contrast, measure per-pixel temporal intensity changes asynchronously. Recovering intensity from events is a popular research topic since the reconstructed images inherit the high dynamic range (HDR) and high-speed properties of events; hence they can be used in many robotic vision applications and to generate slow-motion HDR videos. However, state-of-the-art methods tackle this problem by training an event-to-image recurrent neural network (RNN), which lacks explainability and is difficult to tune. In this work we show, for the first time, how tackling the joint problem of motion and intensity estimation leads us to model event-based image reconstruction as a linear inverse problem that can be solved without training an image reconstruction RNN. Instead, classical and learning-based image priors can be used to solve the problem and remove artifacts from the reconstructed images. The experiments show that the proposed approach generates images with visual quality on par with state-of-the-art methods despite only using data from a short time interval (i.e., without recurrent connections). Our method can also be used to improve the quality of images reconstructed by approaches that first estimate the image Laplacian; here our method can be interpreted as Poisson reconstruction guided by image priors.
Markov-Lipschitz Deep Learning
Jun 21, 2020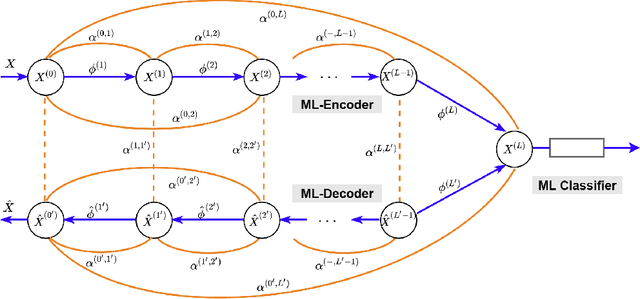



Abstract:In this paper, we propose a novel framework, called Markov-Lipschitz deep learning (MLDL), for manifold learning and data generation. It enhances layer-wise transformations of neural networks by optimizing isometry (geometry-preserving) and stability of the mappings. A prior constraint of locally isometric smoothness (LIS) is imposed across-layers and encoded via a Markov random field (MRF)-Gibbs distribution, and consequently layer-wise vector transformations are replaced by LIS-constrained metric homeomorphisms. These lead to the best possible solutions as measured by locally geometric distortion and locally bi-Lipschitz continuity. Extensive experiments, comparisons and ablation study demonstrate significant advantages of MLDL for manifold learning and data generation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge