Yuzong Chen
P3-LLM: An Integrated NPU-PIM Accelerator for LLM Inference Using Hybrid Numerical Formats
Nov 16, 2025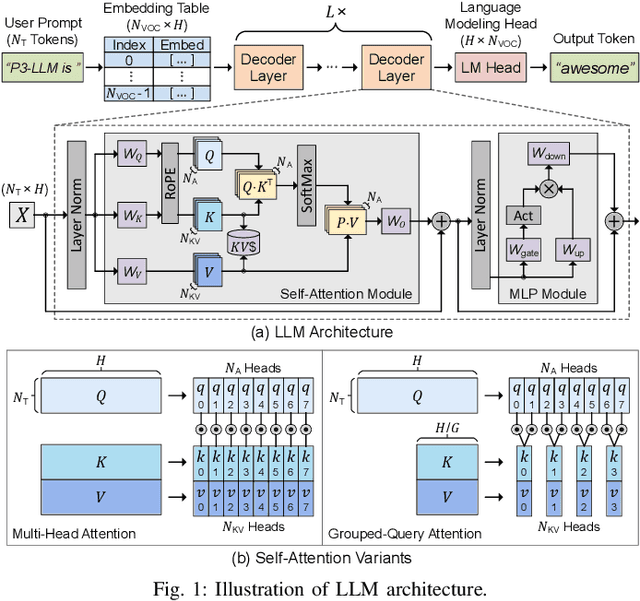
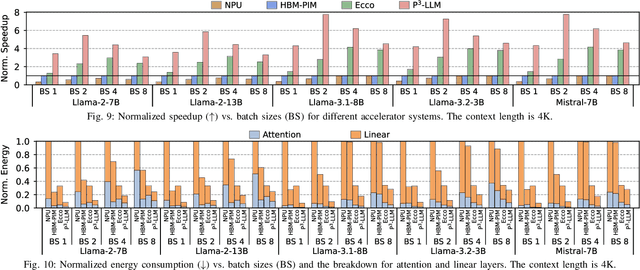
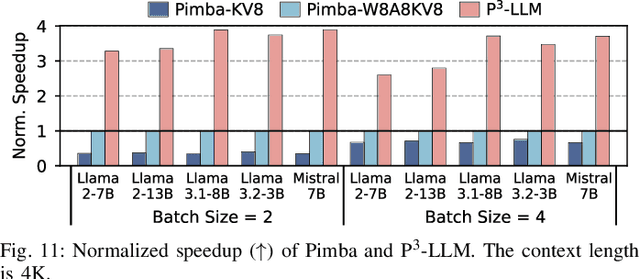
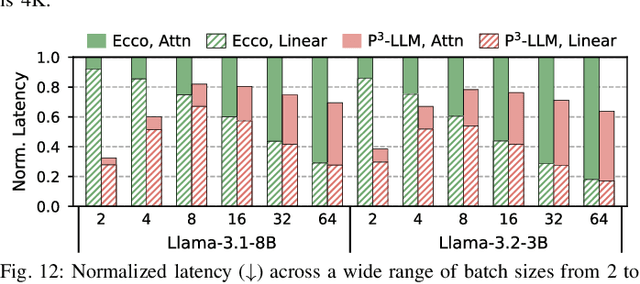
Abstract:The substantial memory bandwidth and computational demands of large language models (LLMs) present critical challenges for efficient inference. To tackle this, the literature has explored heterogeneous systems that combine neural processing units (NPUs) with DRAM-based processing-in-memory (PIM) for LLM acceleration. However, existing high-precision (e.g., FP16) PIM compute units incur significant area and power overhead in DRAM technology, limiting the effective computation throughput. In this paper, we introduce P3-LLM, a novel NPU-PIM integrated accelerator for LLM inference using hybrid numerical formats. Our approach is threefold: First, we propose a flexible mixed-precision quantization scheme, which leverages hybrid numerical formats to quantize different LLM operands with high compression efficiency and minimal accuracy loss. Second, we architect an efficient PIM accelerator for P3-LLM, featuring enhanced compute units to support hybrid numerical formats. Our careful choice of numerical formats allows to co-design low-precision PIM compute units that significantly boost the computation throughput under iso-area constraints. Third, we optimize the low-precision dataflow of different LLM modules by applying operator fusion to minimize the overhead of runtime dequantization. Evaluation on a diverse set of representative LLMs and tasks demonstrates that P3-LLM achieves state-of-the-art accuracy in terms of both KV-cache quantization and weight-activation quantization. Combining the proposed quantization scheme with PIM architecture co-design, P3-LLM yields an average of $4.9\times$, $2.0\times$, and $3.4\times$ speedups over the state-of-the-art LLM accelerators HBM-PIM, Ecco, and Pimba, respectively. Our quantization code is available at https://github.com/yc2367/P3-LLM.git
The Power of Negative Zero: Datatype Customization for Quantized Large Language Models
Jan 06, 2025



Abstract:Large language models (LLMs) have demonstrated remarkable performance across various machine learning tasks, quickly becoming one of the most prevalent AI workloads. Yet the substantial memory requirement of LLMs significantly hinders their deployment for end users. Post-training quantization (PTQ) serves as one of the most hardware-efficient methods to mitigate the memory and computational demands of LLMs. Although the traditional integer (INT) datatype has received widespread adoption in PTQ methods, floating-point (FP) quantization has emerged as a viable alternative thanks to its effectiveness in fitting LLM numerical distributions. However, the FP datatype in sign-magnitude binary representation contains both positive and negative zero, which constrains its representation capability, particularly under low precision (3 and 4 bits). In this paper, we extend the basic FP datatype to perform Redundant Zero Remapping (RaZeR), which remaps the negative zero FP encoding to a set of pre-defined special values to maximally utilize FP quantization encodings and to better fit LLM numerical distributions. Through careful selection of special values, RaZeR outperforms conventional asymmetric INT quantization while achieving high computational efficiency. We demonstrate that RaZeR can be seamlessly integrated with quantization algorithms for both weights and KV-cache, including advanced methods with clipping and transformations, and consistently achieve better model accuracy. Additionally, we implement a fast GEMV kernel with fused dequantization that efficiently converts the 4-bit RaZeR value to FP16 through novel bit-level manipulation. On modern GPUs, our evaluation shows that RaZeR improves the GEMV speed by up to 7.56$\times$ compared to the FP16 implementation, while achieving up to 2.72$\times$ speedup in the LLM decoding throughput.
BitMoD: Bit-serial Mixture-of-Datatype LLM Acceleration
Nov 18, 2024



Abstract:Large language models (LLMs) have demonstrated remarkable performance across various machine learning tasks. Yet the substantial memory footprint of LLMs significantly hinders their deployment. In this paper, we improve the accessibility of LLMs through BitMoD, an algorithm-hardware co-design solution that enables efficient LLM acceleration at low weight precision. On the algorithm side, BitMoD introduces fine-grained data type adaptation that uses a different numerical data type to quantize a group of (e.g., 128) weights. Through the careful design of these new data types, BitMoD is able to quantize LLM weights to very low precision (e.g., 4 bits and 3 bits) while maintaining high accuracy. On the hardware side, BitMoD employs a bit-serial processing element to easily support multiple numerical precisions and data types; our hardware design includes two key innovations: First, it employs a unified representation to process different weight data types, thus reducing the hardware cost. Second, it adopts a bit-serial dequantization unit to rescale the per-group partial sum with minimal hardware overhead. Our evaluation on six representative LLMs demonstrates that BitMoD significantly outperforms state-of-the-art LLM quantization and acceleration methods. For discriminative tasks, BitMoD can quantize LLM weights to 4-bit with $<\!0.5\%$ accuracy loss on average. For generative tasks, BitMoD is able to quantize LLM weights to 3-bit while achieving better perplexity than prior LLM quantization scheme. Combining the superior model performance with an efficient accelerator design, BitMoD achieves an average of $1.69\times$ and $1.48\times$ speedups compared to prior LLM accelerators ANT and OliVe, respectively.
BBS: Bi-directional Bit-level Sparsity for Deep Learning Acceleration
Sep 08, 2024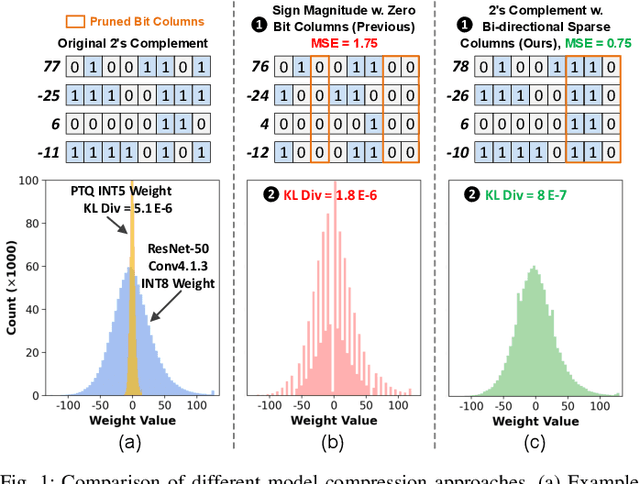
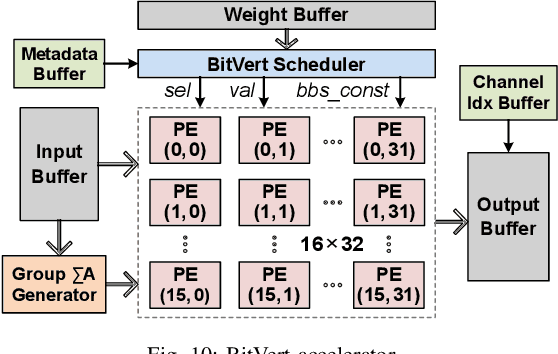
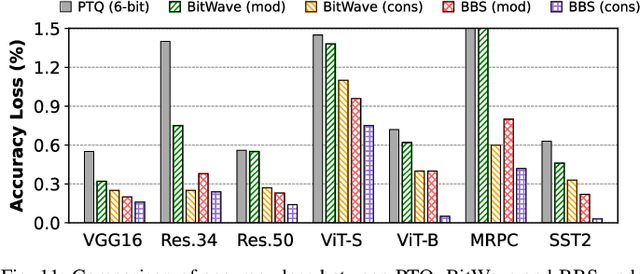
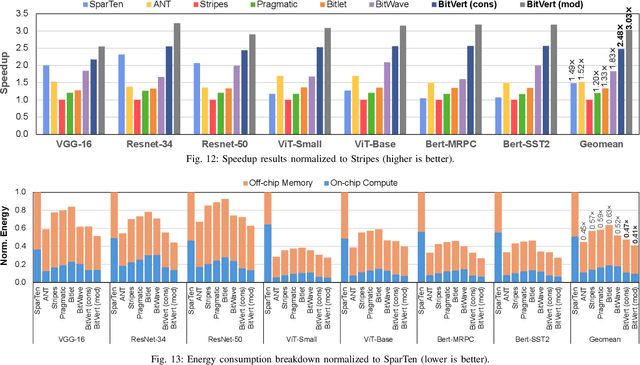
Abstract:Bit-level sparsity methods skip ineffectual zero-bit operations and are typically applicable within bit-serial deep learning accelerators. This type of sparsity at the bit-level is especially interesting because it is both orthogonal and compatible with other deep neural network (DNN) efficiency methods such as quantization and pruning. In this work, we improve the practicality and efficiency of bitlevel sparsity through a novel algorithmic bit-pruning, averaging, and compression method, and a co-designed efficient bit-serial hardware accelerator. On the algorithmic side, we introduce bidirectional bit sparsity (BBS). The key insight of BBS is that we can leverage bit sparsity in a symmetrical way to prune either zero-bits or one-bits. This significantly improves the load balance of bit-serial computing and guarantees the level of sparsity to be more than 50%. On top of BBS, we further propose two bit-level binary pruning methods that require no retraining, and can be seamlessly applied to quantized DNNs. Combining binary pruning with a new tensor encoding scheme, BBS can both skip computation and reduce the memory footprint associated with bi-directional sparse bit columns. On the hardware side, we demonstrate the potential of BBS through BitVert, a bitserial architecture with an efficient PE design to accelerate DNNs with low overhead, exploiting our proposed binary pruning. Evaluation on seven representative DNN models shows that our approach achieves: (1) on average 1.66$\times$ reduction in model sizewith negligible accuracy loss of < 0.5%; (2) up to 3.03$\times$ speedupand 2.44$\times$ energy saving compared to prior DNN accelerators.
Learning from Students: Applying t-Distributions to Explore Accurate and Efficient Formats for LLMs
May 06, 2024



Abstract:Large language models (LLMs) have recently achieved state-of-the-art performance across various tasks, yet due to their large computational requirements, they struggle with strict latency and power demands. Deep neural network (DNN) quantization has traditionally addressed these limitations by converting models to low-precision integer formats. Yet recently alternative formats, such as Normal Float (NF4), have been shown to consistently increase model accuracy, albeit at the cost of increased chip area. In this work, we first conduct a large-scale analysis of LLM weights and activations across 30 networks to conclude most distributions follow a Student's t-distribution. We then derive a new theoretically optimal format, Student Float (SF4), with respect to this distribution, that improves over NF4 across modern LLMs, for example increasing the average accuracy on LLaMA2-7B by 0.76% across tasks. Using this format as a high-accuracy reference, we then propose augmenting E2M1 with two variants of supernormal support for higher model accuracy. Finally, we explore the quality and performance frontier across 11 datatypes, including non-traditional formats like Additive-Powers-of-Two (APoT), by evaluating their model accuracy and hardware complexity. We discover a Pareto curve composed of INT4, E2M1, and E2M1 with supernormal support, which offers a continuous tradeoff between model accuracy and chip area. For example, E2M1 with supernormal support increases the accuracy of Phi-2 by up to 2.19% with 1.22% area overhead, enabling more LLM-based applications to be run at four bits.
Clustering Bioactive Molecules in 3D Chemical Space with Unsupervised Deep Learning
Feb 09, 2019Abstract:Unsupervised clustering has broad applications in data stratification, pattern investigation and new discovery beyond existing knowledge. In particular, clustering of bioactive molecules facilitates chemical space mapping, structure-activity studies, and drug discovery. These tasks, conventionally conducted by similarity-based methods, are complicated by data complexity and diversity. We ex-plored the superior learning capability of deep autoencoders for unsupervised clustering of 1.39 mil-lion bioactive molecules into band-clusters in a 3-dimensional latent chemical space. These band-clusters, displayed by a space-navigation simulation software, band molecules of selected bioactivity classes into individual band-clusters possessing unique sets of common sub-structural features beyond structural similarity. These sub-structural features form the frameworks of the literature-reported pharmacophores and privileged fragments. Within each band-cluster, molecules are further banded into selected sub-regions with respect to their bioactivity target, sub-structural features and molecular scaffolds. Our method is potentially applicable for big data clustering tasks of different fields.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge