Xavier Llado
Benchmarking the CoW with the TopCoW Challenge: Topology-Aware Anatomical Segmentation of the Circle of Willis for CTA and MRA
Dec 29, 2023



Abstract:The Circle of Willis (CoW) is an important network of arteries connecting major circulations of the brain. Its vascular architecture is believed to affect the risk, severity, and clinical outcome of serious neuro-vascular diseases. However, characterizing the highly variable CoW anatomy is still a manual and time-consuming expert task. The CoW is usually imaged by two angiographic imaging modalities, magnetic resonance angiography (MRA) and computed tomography angiography (CTA), but there exist limited public datasets with annotations on CoW anatomy, especially for CTA. Therefore we organized the TopCoW Challenge in 2023 with the release of an annotated CoW dataset and invited submissions worldwide for the CoW segmentation task, which attracted over 140 registered participants from four continents. TopCoW dataset was the first public dataset with voxel-level annotations for CoW's 13 vessel components, made possible by virtual-reality (VR) technology. It was also the first dataset with paired MRA and CTA from the same patients. TopCoW challenge aimed to tackle the CoW characterization problem as a multiclass anatomical segmentation task with an emphasis on topological metrics. The top performing teams managed to segment many CoW components to Dice scores around 90%, but with lower scores for communicating arteries and rare variants. There were also topological mistakes for predictions with high Dice scores. Additional topological analysis revealed further areas for improvement in detecting certain CoW components and matching CoW variant's topology accurately. TopCoW represented a first attempt at benchmarking the CoW anatomical segmentation task for MRA and CTA, both morphologically and topologically.
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021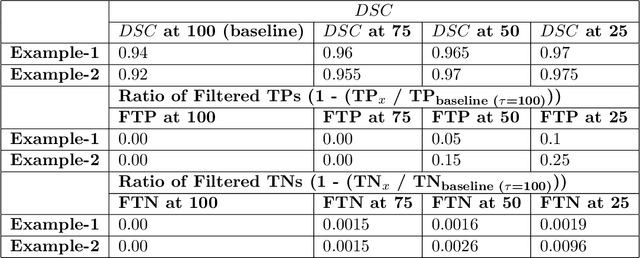
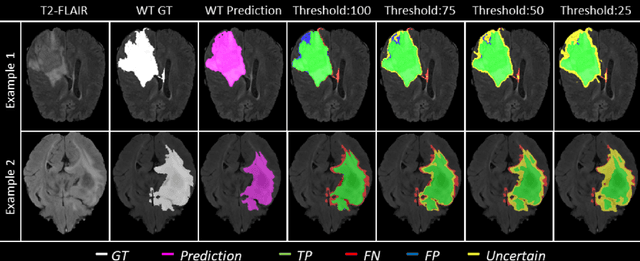
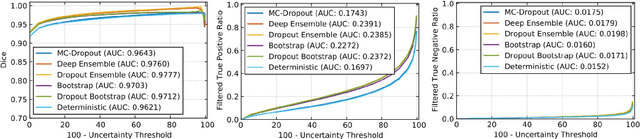
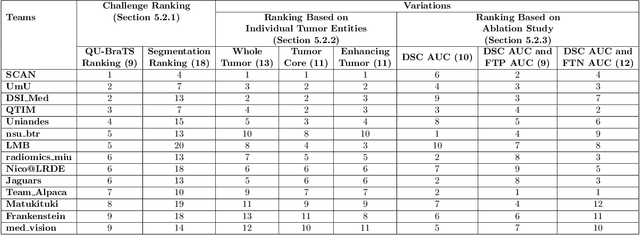
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
Automated sub-cortical brain structure segmentation combining spatial and deep convolutional features
Sep 26, 2017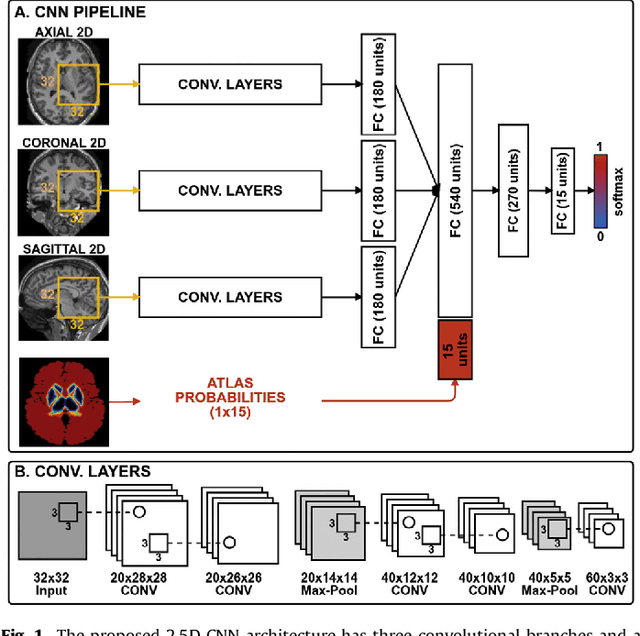
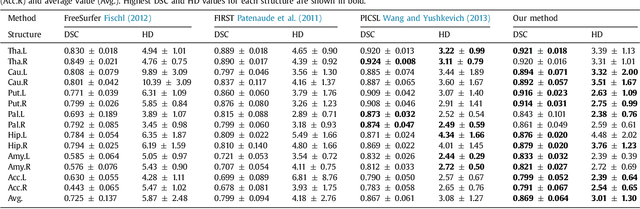
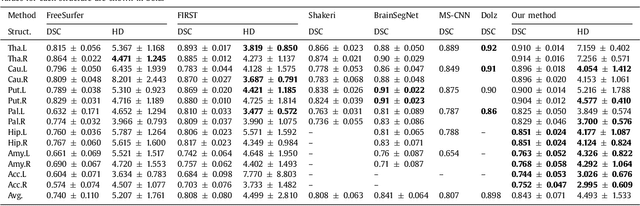
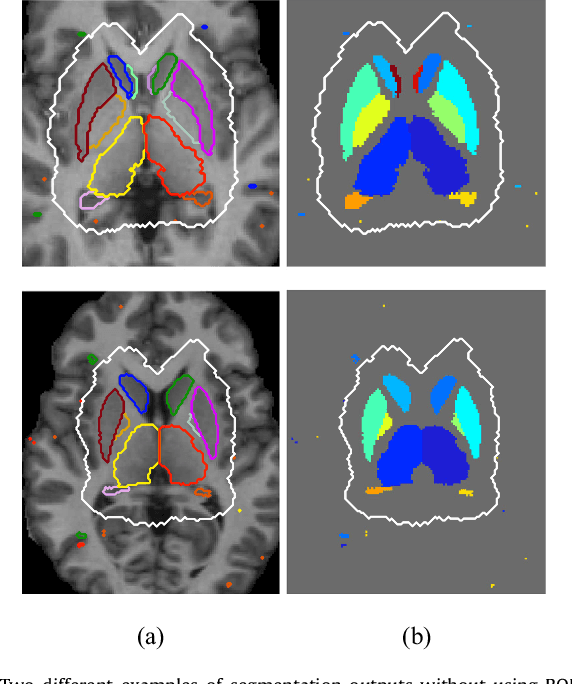
Abstract:Sub-cortical brain structure segmentation in Magnetic Resonance Images (MRI) has attracted the interest of the research community for a long time because morphological changes in these structures are related to different neurodegenerative disorders. However, manual segmentation of these structures can be tedious and prone to variability, highlighting the need for robust automated segmentation methods. In this paper, we present a novel convolutional neural network based approach for accurate segmentation of the sub-cortical brain structures that combines both convolutional and prior spatial features for improving the segmentation accuracy. In order to increase the accuracy of the automated segmentation, we propose to train the network using a restricted sample selection to force the network to learn the most difficult parts of the structures. We evaluate the accuracy of the proposed method on the public MICCAI 2012 challenge and IBSR 18 datasets, comparing it with different available state-of-the-art methods and other recently proposed deep learning approaches. On the MICCAI 2012 dataset, our method shows an excellent performance comparable to the best challenge participant strategy, while performing significantly better than state-of-the-art techniques such as FreeSurfer and FIRST. On the IBSR 18 dataset, our method also exhibits a significant increase in the performance with respect to not only FreeSurfer and FIRST, but also comparable or better results than other recent deep learning approaches. Moreover, our experiments show that both the addition of the spatial priors and the restricted sampling strategy have a significant effect on the accuracy of the proposed method. In order to encourage the reproducibility and the use of the proposed method, a public version of our approach is available to download for the neuroimaging community.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge