Vithya Yogarajan
Tackling Bias in Pre-trained Language Models: Current Trends and Under-represented Societies
Dec 03, 2023Abstract:The benefits and capabilities of pre-trained language models (LLMs) in current and future innovations are vital to any society. However, introducing and using LLMs comes with biases and discrimination, resulting in concerns about equality, diversity and fairness, and must be addressed. While understanding and acknowledging bias in LLMs and developing mitigation strategies are crucial, the generalised assumptions towards societal needs can result in disadvantages towards under-represented societies and indigenous populations. Furthermore, the ongoing changes to actual and proposed amendments to regulations and laws worldwide also impact research capabilities in tackling the bias problem. This research presents a comprehensive survey synthesising the current trends and limitations in techniques used for identifying and mitigating bias in LLMs, where the overview of methods for tackling bias are grouped into metrics, benchmark datasets, and mitigation strategies. The importance and novelty of this survey are that it explores the perspective of under-represented societies. We argue that current practices tackling the bias problem cannot simply be 'plugged in' to address the needs of under-represented societies. We use examples from New Zealand to present requirements for adopting existing techniques to under-represented societies.
Challenges in Annotating Datasets to Quantify Bias in Under-represented Society
Sep 11, 2023
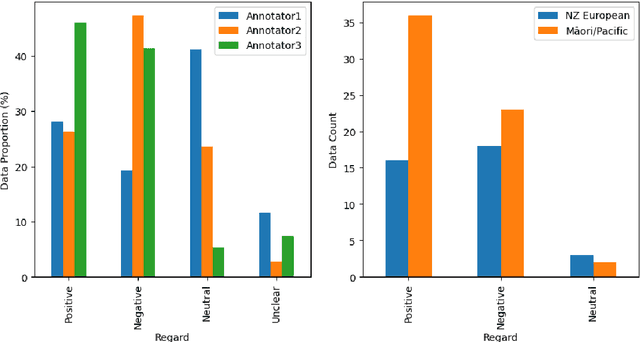


Abstract:Recent advances in artificial intelligence, including the development of highly sophisticated large language models (LLM), have proven beneficial in many real-world applications. However, evidence of inherent bias encoded in these LLMs has raised concerns about equity. In response, there has been an increase in research dealing with bias, including studies focusing on quantifying bias and developing debiasing techniques. Benchmark bias datasets have also been developed for binary gender classification and ethical/racial considerations, focusing predominantly on American demographics. However, there is minimal research in understanding and quantifying bias related to under-represented societies. Motivated by the lack of annotated datasets for quantifying bias in under-represented societies, we endeavoured to create benchmark datasets for the New Zealand (NZ) population. We faced many challenges in this process, despite the availability of three annotators. This research outlines the manual annotation process, provides an overview of the challenges we encountered and lessons learnt, and presents recommendations for future research.
Neuromodulation Gated Transformer
May 11, 2023Abstract:We introduce a novel architecture, the Neuromodulation Gated Transformer (NGT), which is a simple implementation of neuromodulation in transformers via a multiplicative effect. We compare it to baselines and show that it results in the best average performance on the SuperGLUE benchmark validation sets.
Effectiveness of Debiasing Techniques: An Indigenous Qualitative Analysis
Apr 17, 2023Abstract:An indigenous perspective on the effectiveness of debiasing techniques for pre-trained language models (PLMs) is presented in this paper. The current techniques used to measure and debias PLMs are skewed towards the US racial biases and rely on pre-defined bias attributes (e.g. "black" vs "white"). Some require large datasets and further pre-training. Such techniques are not designed to capture the underrepresented indigenous populations in other countries, such as M\=aori in New Zealand. Local knowledge and understanding must be incorporated to ensure unbiased algorithms, especially when addressing a resource-restricted society.
* accepted with invite to present
The Development of a Labelled te reo Māori-English Bilingual Database for Language Technology
Aug 21, 2022



Abstract:Te reo M\=aori (referred to as M\=aori), New Zealand's indigenous language, is under-resourced in language technology. M\=aori speakers are bilingual, where M\=aori is code-switched with English. Unfortunately, there are minimal resources available for M\=aori language technology, language detection and code-switch detection between M\=aori-English pair. Both English and M\=aori use Roman-derived orthography making rule-based systems for detecting language and code-switching restrictive. Most M\=aori language detection is done manually by language experts. This research builds a M\=aori-English bilingual database of 66,016,807 words with word-level language annotation. The New Zealand Parliament Hansard debates reports were used to build the database. The language labels are assigned using language-specific rules and expert manual annotations. Words with the same spelling, but different meanings, exist for M\=aori and English. These words could not be categorised as M\=aori or English based on word-level language rules. Hence, manual annotations were necessary. An analysis reporting the various aspects of the database such as metadata, year-wise analysis, frequently occurring words, sentence length and N-grams is also reported. The database developed here is a valuable tool for future language and speech technology development for Aotearoa New Zealand. The methodology followed to label the database can also be followed by other low-resourced language pairs.
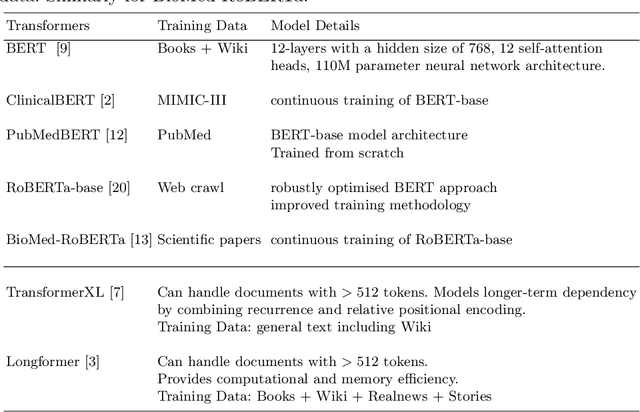
Improving Predictions of Tail-end Labels using Concatenated BioMed-Transformers for Long Medical Documents
Dec 03, 2021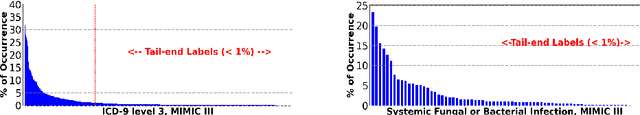
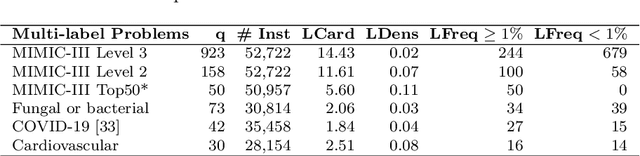
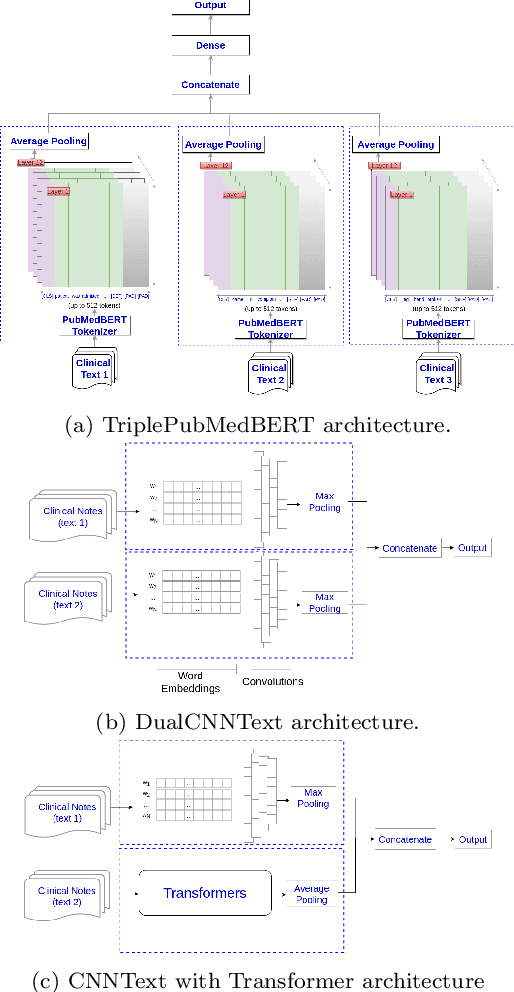
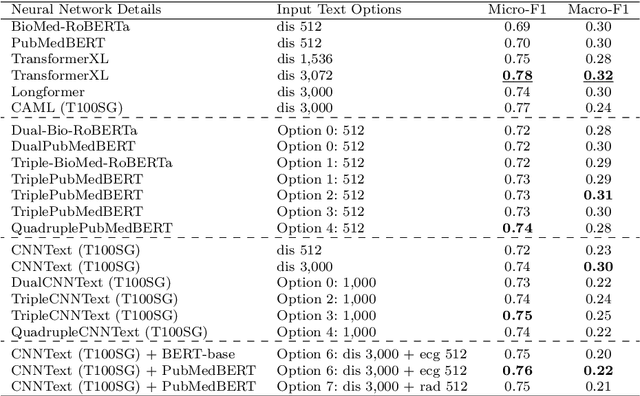
Abstract:Multi-label learning predicts a subset of labels from a given label set for an unseen instance while considering label correlations. A known challenge with multi-label classification is the long-tailed distribution of labels. Many studies focus on improving the overall predictions of the model and thus do not prioritise tail-end labels. Improving the tail-end label predictions in multi-label classifications of medical text enables the potential to understand patients better and improve care. The knowledge gained by one or more infrequent labels can impact the cause of medical decisions and treatment plans. This research presents variations of concatenated domain-specific language models, including multi-BioMed-Transformers, to achieve two primary goals. First, to improve F1 scores of infrequent labels across multi-label problems, especially with long-tail labels; second, to handle long medical text and multi-sourced electronic health records (EHRs), a challenging task for standard transformers designed to work on short input sequences. A vital contribution of this research is new state-of-the-art (SOTA) results obtained using TransformerXL for predicting medical codes. A variety of experiments are performed on the Medical Information Mart for Intensive Care (MIMIC-III) database. Results show that concatenated BioMed-Transformers outperform standard transformers in terms of overall micro and macro F1 scores and individual F1 scores of tail-end labels, while incurring lower training times than existing transformer-based solutions for long input sequences.
Predicting COVID-19 Patient Shielding: A Comprehensive Study
Oct 01, 2021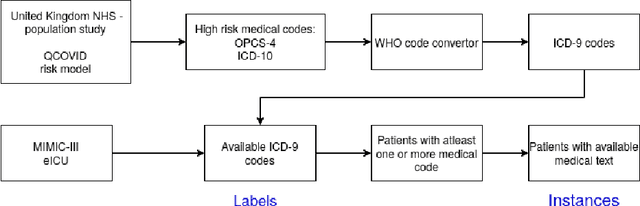
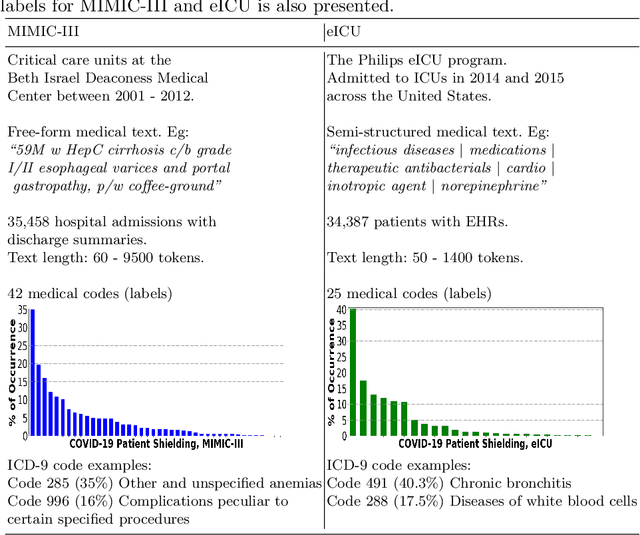
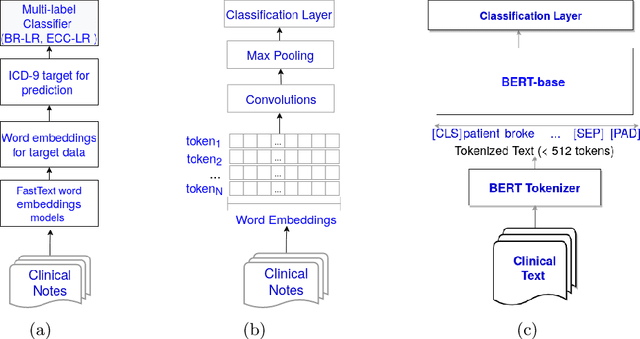
Abstract:There are many ways machine learning and big data analytics are used in the fight against the COVID-19 pandemic, including predictions, risk management, diagnostics, and prevention. This study focuses on predicting COVID-19 patient shielding -- identifying and protecting patients who are clinically extremely vulnerable from coronavirus. This study focuses on techniques used for the multi-label classification of medical text. Using the information published by the United Kingdom NHS and the World Health Organisation, we present a novel approach to predicting COVID-19 patient shielding as a multi-label classification problem. We use publicly available, de-identified ICU medical text data for our experiments. The labels are derived from the published COVID-19 patient shielding data. We present an extensive comparison across 12 multi-label classifiers from the simple binary relevance to neural networks and the most recent transformers. To the best of our knowledge this is the first comprehensive study, where such a range of multi-label classifiers for medical text are considered. We highlight the benefits of various approaches, and argue that, for the task at hand, both predictive accuracy and processing time are essential.
* Accepted in AJCAI 2021
Seeing The Whole Patient: Using Multi-Label Medical Text Classification Techniques to Enhance Predictions of Medical Codes
Mar 29, 2020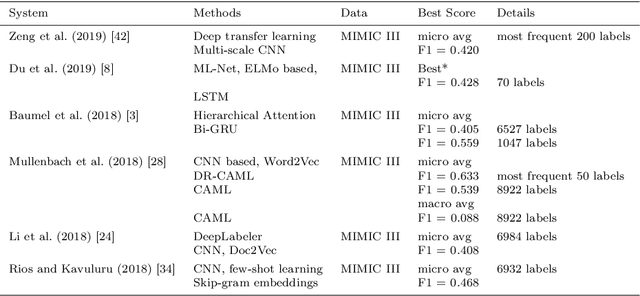
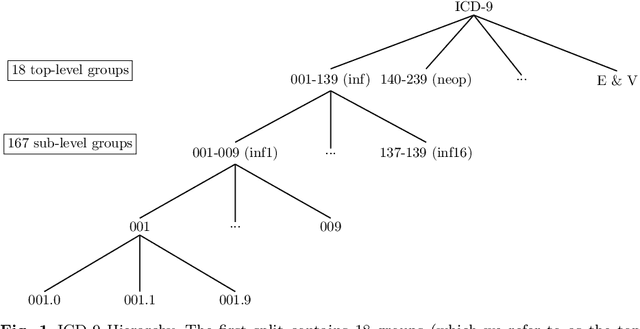
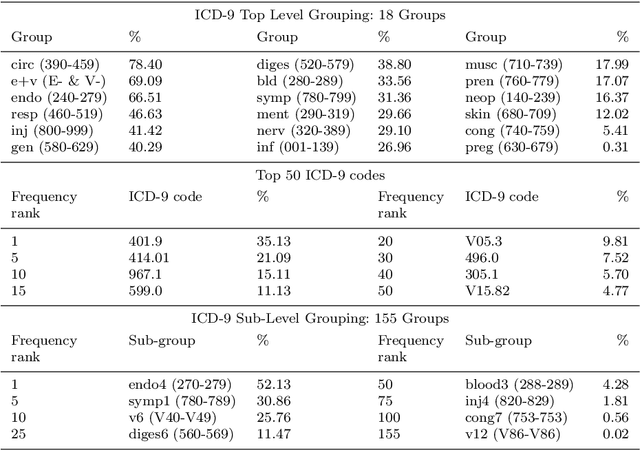
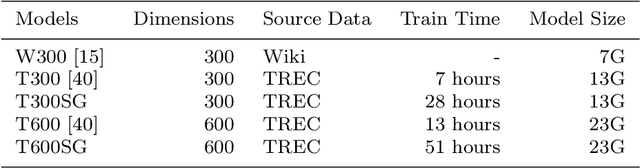
Abstract:Machine learning-based multi-label medical text classifications can be used to enhance the understanding of the human body and aid the need for patient care. We present a broad study on clinical natural language processing techniques to maximise a feature representing text when predicting medical codes on patients with multi-morbidity. We present results of multi-label medical text classification problems with 18, 50 and 155 labels. We compare several variations to embeddings, text tagging, and pre-processing. For imbalanced data we show that labels which occur infrequently, benefit the most from additional features incorporated in embeddings. We also show that high dimensional embeddings pre-trained using health-related data present a significant improvement in a multi-label setting, similarly to the way they improve performance for binary classification. High dimensional embeddings from this research are made available for public use.
Automatic end-to-end De-identification: Is high accuracy the only metric?
Jan 27, 2019



Abstract:De-identification of electronic health records (EHR) is a vital step towards advancing health informatics research and maximising the use of available data. It is a two-step process where step one is the identification of protected health information (PHI), and step two is replacing such PHI with surrogates. Despite the recent advances in automatic de-identification of EHR, significant obstacles remain if the abundant health data available are to be used to the full potential. Accuracy in de-identification could be considered a necessary, but not sufficient condition for the use of EHR without individual patient consent. We present here a comprehensive review of the progress to date, both the impressive successes in achieving high accuracy and the significant risks and challenges that remain. To best of our knowledge, this is the first paper to present a complete picture of end-to-end automatic de-identification. We review 18 recently published automatic de-identification systems -designed to de-identify EHR in the form of free text- to show the advancements made in improving the overall accuracy of the system, and in identifying individual PHI. We argue that despite the improvements in accuracy there remain challenges in surrogate generation and replacements of identified PHIs, and the risks posed to patient protection and privacy.
A survey of automatic de-identification of longitudinal clinical narratives
Oct 16, 2018
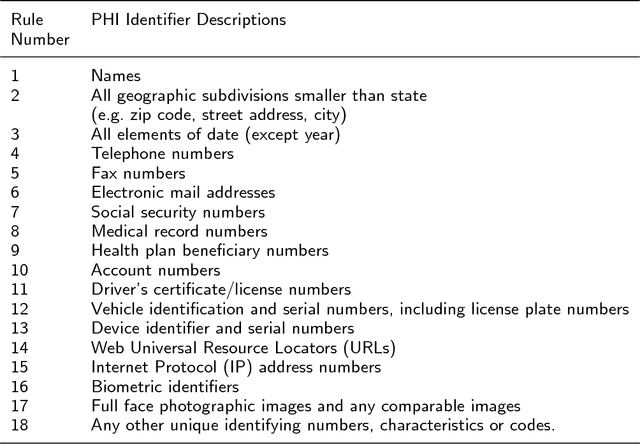
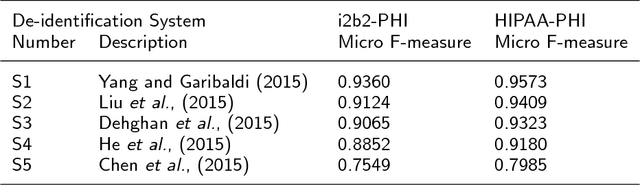
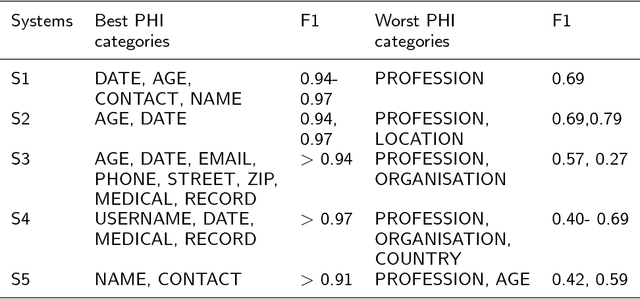
Abstract:Use of medical data, also known as electronic health records, in research helps develop and advance medical science. However, protecting patient confidentiality and identity while using medical data for analysis is crucial. Medical data can be in the form of tabular structures (i.e. tables), free-form narratives, and images. This study focuses on medical data in the free form longitudinal text. De-identification of electronic health records provides the opportunity to use such data for research without it affecting patient privacy, and avoids the need for individual patient consent. In recent years there is increasing interest in developing an accurate, robust and adaptable automatic de-identification system for electronic health records. This is mainly due to the dilemma between the availability of an abundance of health data, and the inability to use such data in research due to legal and ethical restrictions. De-identification tracks in competitions such as the 2014 i2b2 UTHealth and the 2016 CEGS N-GRID shared tasks have provided a great platform to advance this area. The primary reasons for this include the open source nature of the dataset and the fact that raw psychiatric data were used for 2016 competitions. This study focuses on noticeable trend changes in the techniques used in the development of automatic de-identification for longitudinal clinical narratives. More specifically, the shift from using conditional random fields (CRF) based systems only or rules (regular expressions, dictionary or combinations) based systems only, to hybrid models (combining CRF and rules), and more recently to deep learning based systems. We review the literature and results that arose from the 2014 and the 2016 competitions and discuss the outcomes of these systems. We also provide a list of research questions that emerged from this survey.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge