Jacob Montiel
Improving Predictions of Tail-end Labels using Concatenated BioMed-Transformers for Long Medical Documents
Dec 03, 2021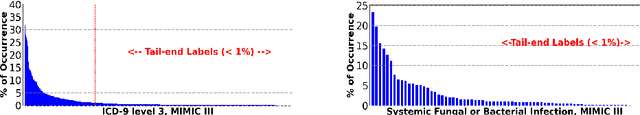
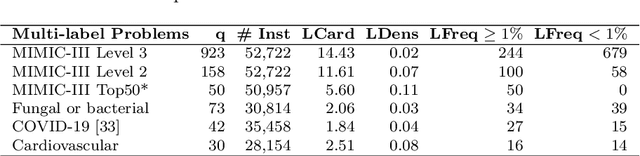
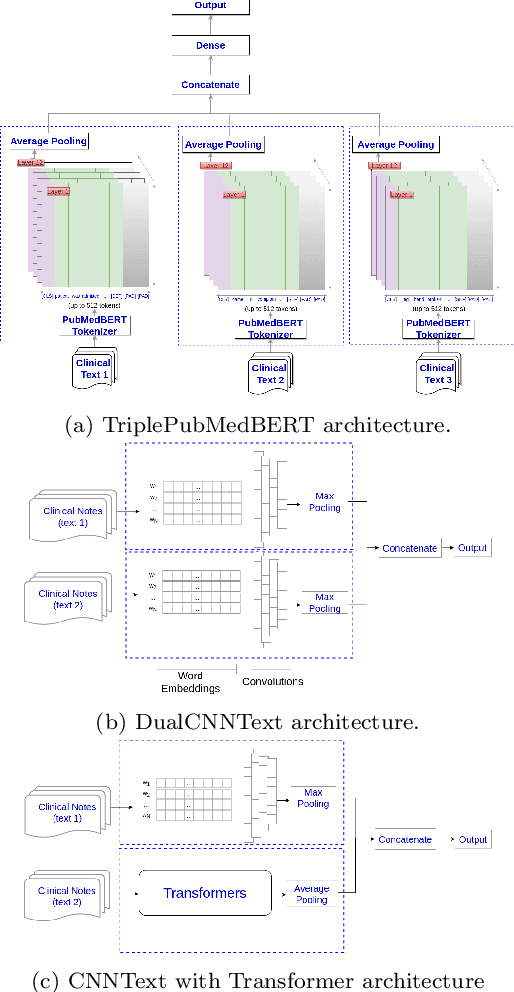
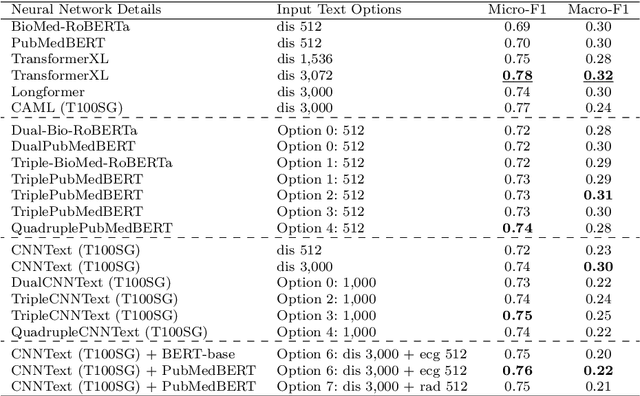
Abstract:Multi-label learning predicts a subset of labels from a given label set for an unseen instance while considering label correlations. A known challenge with multi-label classification is the long-tailed distribution of labels. Many studies focus on improving the overall predictions of the model and thus do not prioritise tail-end labels. Improving the tail-end label predictions in multi-label classifications of medical text enables the potential to understand patients better and improve care. The knowledge gained by one or more infrequent labels can impact the cause of medical decisions and treatment plans. This research presents variations of concatenated domain-specific language models, including multi-BioMed-Transformers, to achieve two primary goals. First, to improve F1 scores of infrequent labels across multi-label problems, especially with long-tail labels; second, to handle long medical text and multi-sourced electronic health records (EHRs), a challenging task for standard transformers designed to work on short input sequences. A vital contribution of this research is new state-of-the-art (SOTA) results obtained using TransformerXL for predicting medical codes. A variety of experiments are performed on the Medical Information Mart for Intensive Care (MIMIC-III) database. Results show that concatenated BioMed-Transformers outperform standard transformers in terms of overall micro and macro F1 scores and individual F1 scores of tail-end labels, while incurring lower training times than existing transformer-based solutions for long input sequences.
Predicting COVID-19 Patient Shielding: A Comprehensive Study
Oct 01, 2021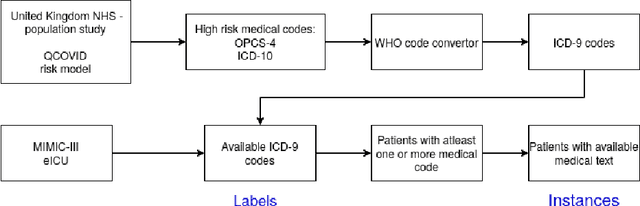
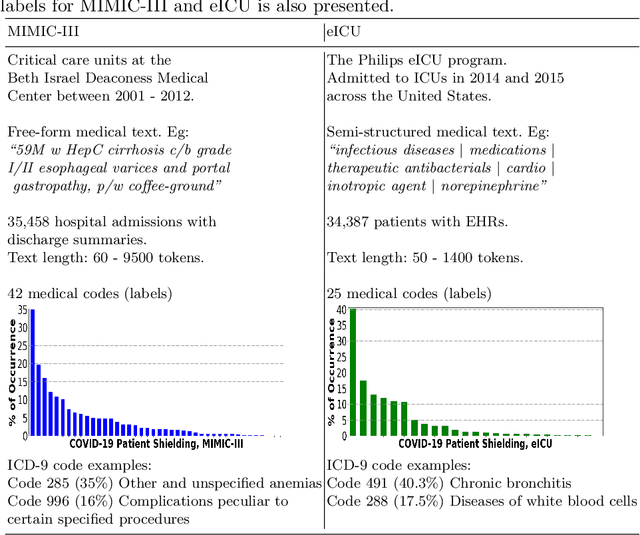
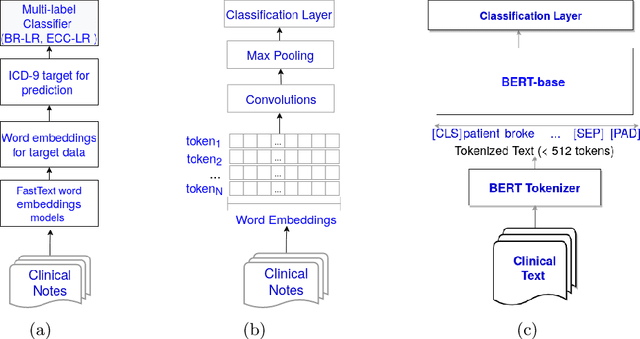
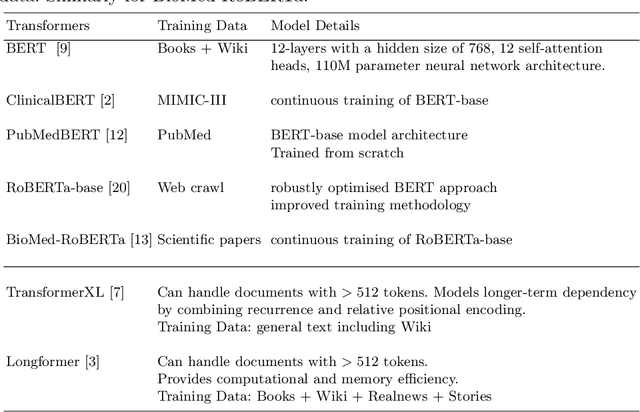
Abstract:There are many ways machine learning and big data analytics are used in the fight against the COVID-19 pandemic, including predictions, risk management, diagnostics, and prevention. This study focuses on predicting COVID-19 patient shielding -- identifying and protecting patients who are clinically extremely vulnerable from coronavirus. This study focuses on techniques used for the multi-label classification of medical text. Using the information published by the United Kingdom NHS and the World Health Organisation, we present a novel approach to predicting COVID-19 patient shielding as a multi-label classification problem. We use publicly available, de-identified ICU medical text data for our experiments. The labels are derived from the published COVID-19 patient shielding data. We present an extensive comparison across 12 multi-label classifiers from the simple binary relevance to neural networks and the most recent transformers. To the best of our knowledge this is the first comprehensive study, where such a range of multi-label classifiers for medical text are considered. We highlight the benefits of various approaches, and argue that, for the task at hand, both predictive accuracy and processing time are essential.
* Accepted in AJCAI 2021
River: machine learning for streaming data in Python
Dec 08, 2020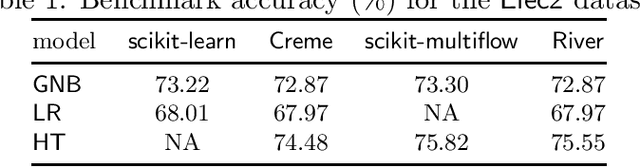
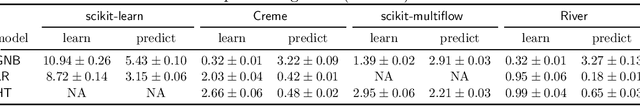
Abstract:River is a machine learning library for dynamic data streams and continual learning. It provides multiple state-of-the-art learning methods, data generators/transformers, performance metrics and evaluators for different stream learning problems. It is the result from the merger of the two most popular packages for stream learning in Python: Creme and scikit-multiflow. River introduces a revamped architecture based on the lessons learnt from the seminal packages. River's ambition is to be the go-to library for doing machine learning on streaming data. Additionally, this open source package brings under the same umbrella a large community of practitioners and researchers. The source code is available at https://github.com/online-ml/river.
Adaptive XGBoost for Evolving Data Streams
May 15, 2020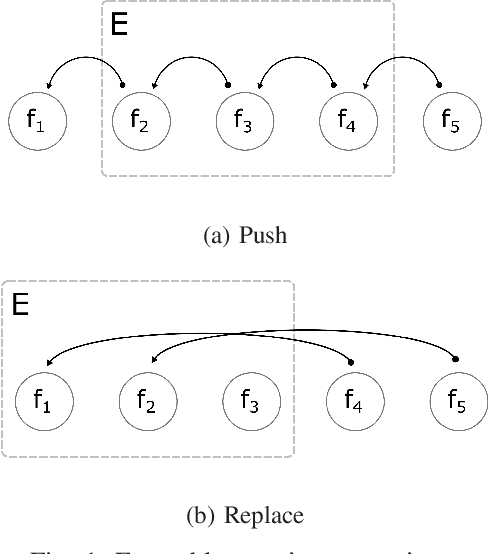
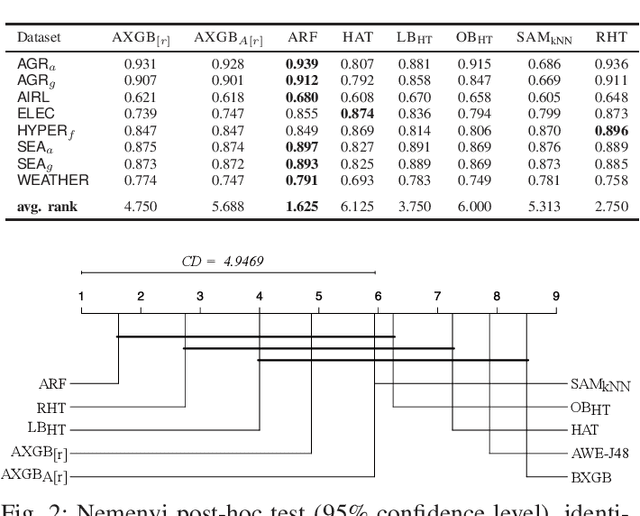
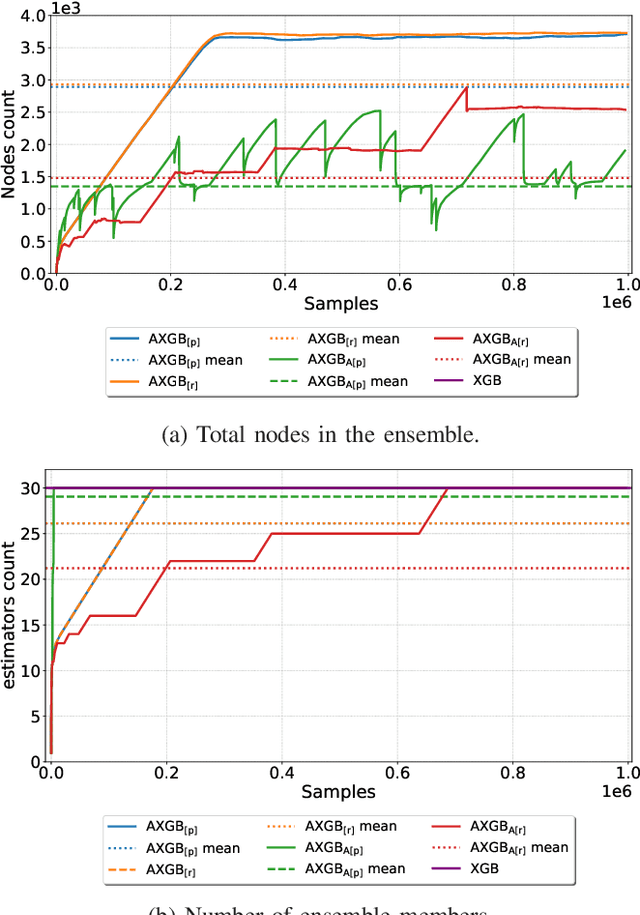
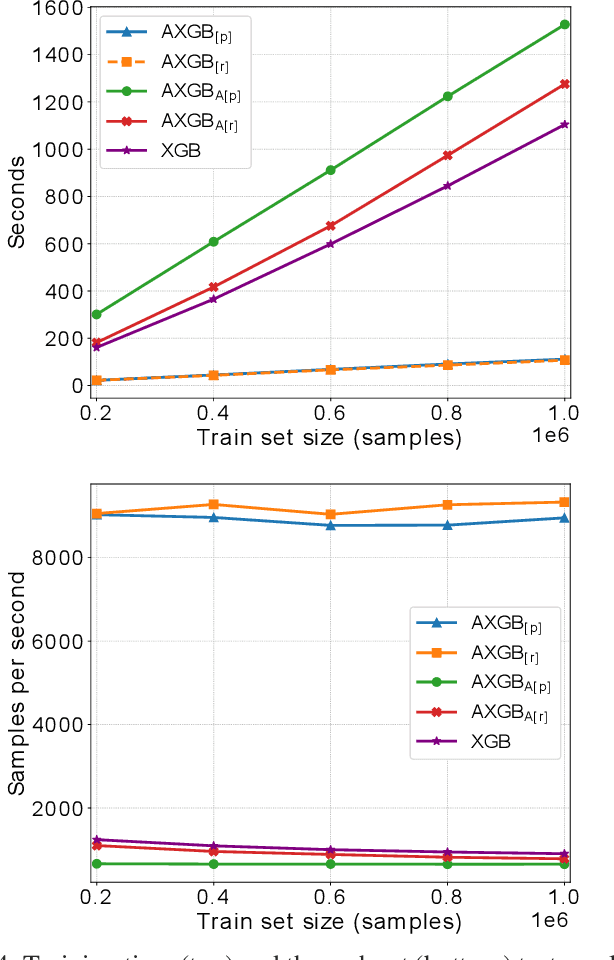
Abstract:Boosting is an ensemble method that combines base models in a sequential manner to achieve high predictive accuracy. A popular learning algorithm based on this ensemble method is eXtreme Gradient Boosting (XGB). We present an adaptation of XGB for classification of evolving data streams. In this setting, new data arrives over time and the relationship between the class and the features may change in the process, thus exhibiting concept drift. The proposed method creates new members of the ensemble from mini-batches of data as new data becomes available. The maximum ensemble size is fixed, but learning does not stop when this size is reached because the ensemble is updated on new data to ensure consistency with the current concept. We also explore the use of concept drift detection to trigger a mechanism to update the ensemble. We test our method on real and synthetic data with concept drift and compare it against batch-incremental and instance-incremental classification methods for data streams.
Seeing The Whole Patient: Using Multi-Label Medical Text Classification Techniques to Enhance Predictions of Medical Codes
Mar 29, 2020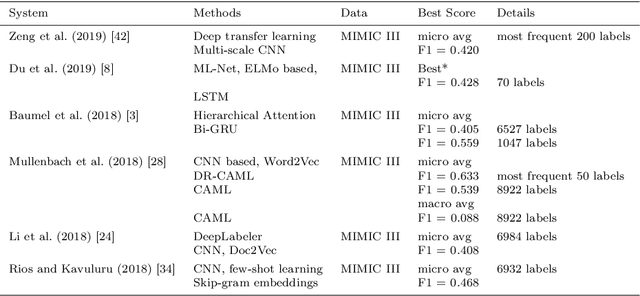
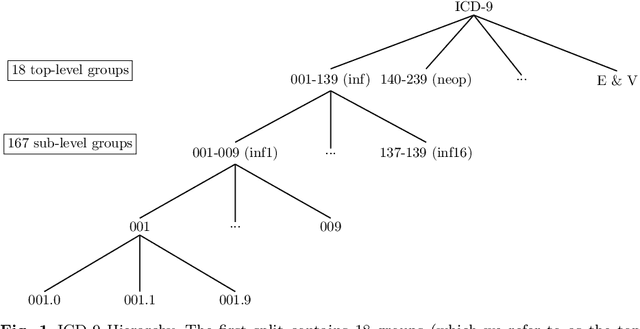
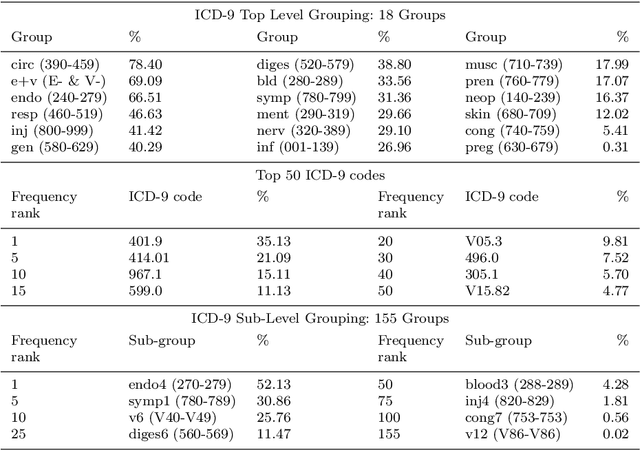
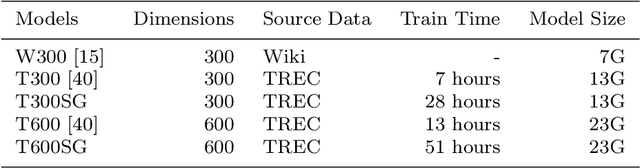
Abstract:Machine learning-based multi-label medical text classifications can be used to enhance the understanding of the human body and aid the need for patient care. We present a broad study on clinical natural language processing techniques to maximise a feature representing text when predicting medical codes on patients with multi-morbidity. We present results of multi-label medical text classification problems with 18, 50 and 155 labels. We compare several variations to embeddings, text tagging, and pre-processing. For imbalanced data we show that labels which occur infrequently, benefit the most from additional features incorporated in embeddings. We also show that high dimensional embeddings pre-trained using health-related data present a significant improvement in a multi-label setting, similarly to the way they improve performance for binary classification. High dimensional embeddings from this research are made available for public use.
Scikit-Multiflow: A Multi-output Streaming Framework
Jul 12, 2018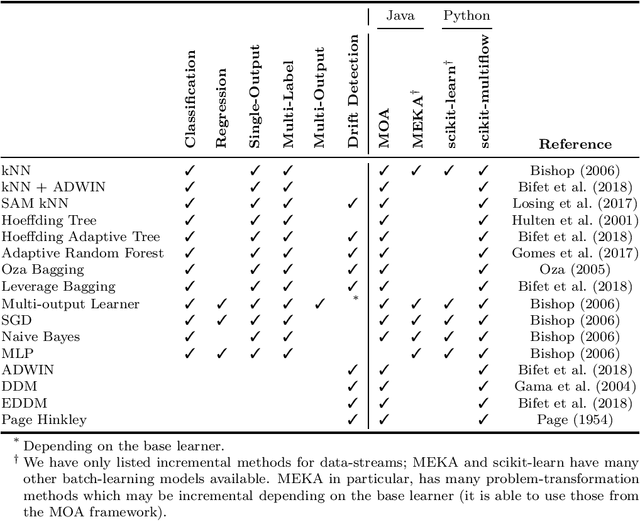
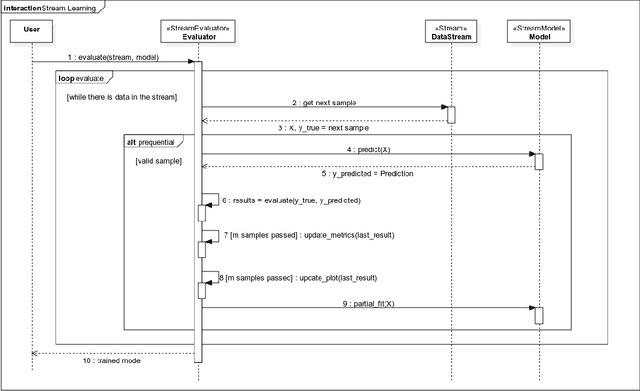
Abstract:Scikit-multiflow is a multi-output/multi-label and stream data mining framework for the Python programming language. Conceived to serve as a platform to encourage democratization of stream learning research, it provides multiple state of the art methods for stream learning, stream generators and evaluators. scikit-multiflow builds upon popular open source frameworks including scikit-learn, MOA and MEKA. Development follows the FOSS principles and quality is enforced by complying with PEP8 guidelines and using continuous integration and automatic testing. The source code is publicly available at https://github.com/scikit-multiflow/scikit-multiflow.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge