Tony Smith
Fake News Classification in Urdu: A Domain Adaptation Approach for a Low-Resource Language
Dec 28, 2025Abstract:Misinformation on social media is a widely acknowledged issue, and researchers worldwide are actively engaged in its detection. However, low-resource languages such as Urdu have received limited attention in this domain. An obvious approach is to utilize a multilingual pretrained language model and fine-tune it for a downstream classification task, such as misinformation detection. However, these models struggle with domain-specific terms, leading to suboptimal performance. To address this, we investigate the effectiveness of domain adaptation before fine-tuning for fake news classification in Urdu, employing a staged training approach to optimize model generalization. We evaluate two widely used multilingual models, XLM-RoBERTa and mBERT, and apply domain-adaptive pretraining using a publicly available Urdu news corpus. Experiments on four publicly available Urdu fake news datasets show that domain-adapted XLM-R consistently outperforms its vanilla counterpart, while domain-adapted mBERT exhibits mixed results.
Detection of Human and Machine-Authored Fake News in Urdu
Oct 25, 2024Abstract:The rise of social media has amplified the spread of fake news, now further complicated by large language models (LLMs) like ChatGPT, which ease the generation of highly convincing, error-free misinformation, making it increasingly challenging for the public to discern truth from falsehood. Traditional fake news detection methods relying on linguistic cues also becomes less effective. Moreover, current detectors primarily focus on binary classification and English texts, often overlooking the distinction between machine-generated true vs. fake news and the detection in low-resource languages. To this end, we updated detection schema to include machine-generated news with focus on the Urdu language. We further propose a hierarchical detection strategy to improve the accuracy and robustness. Experiments show its effectiveness across four datasets in various settings.
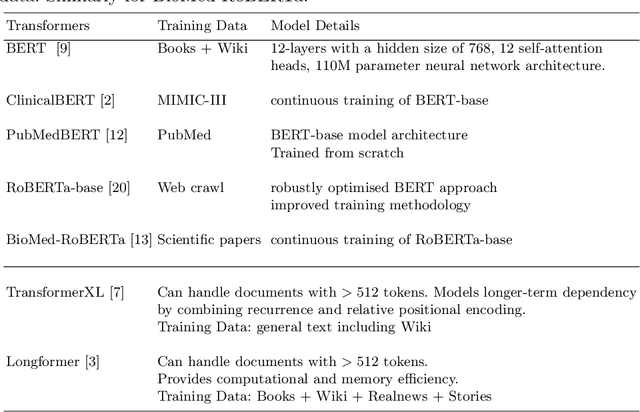
Improving Predictions of Tail-end Labels using Concatenated BioMed-Transformers for Long Medical Documents
Dec 03, 2021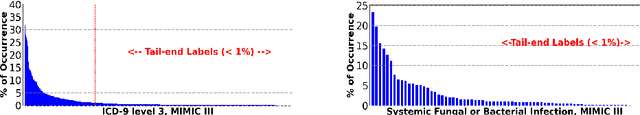
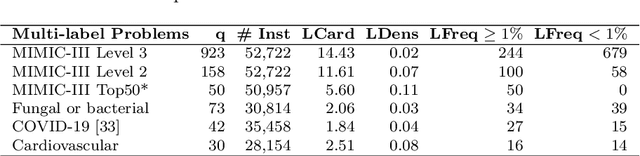
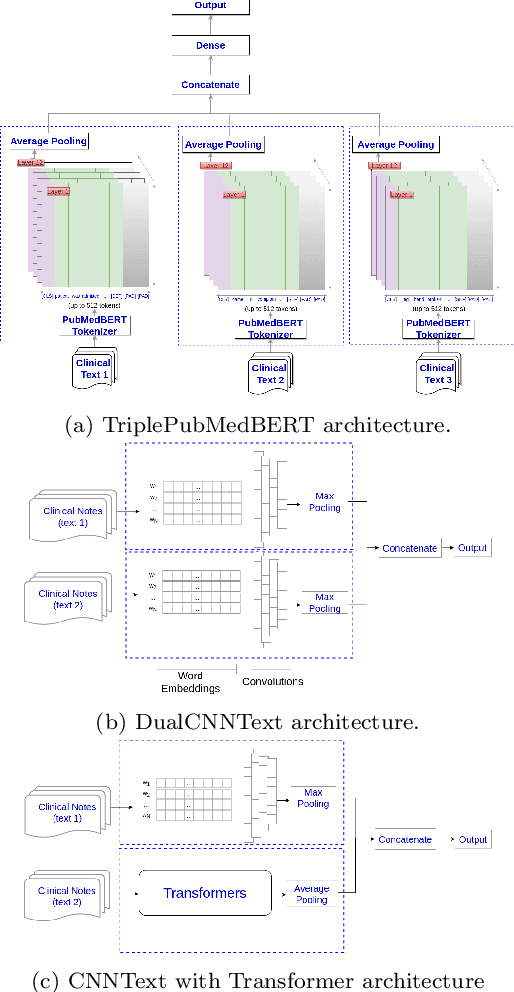
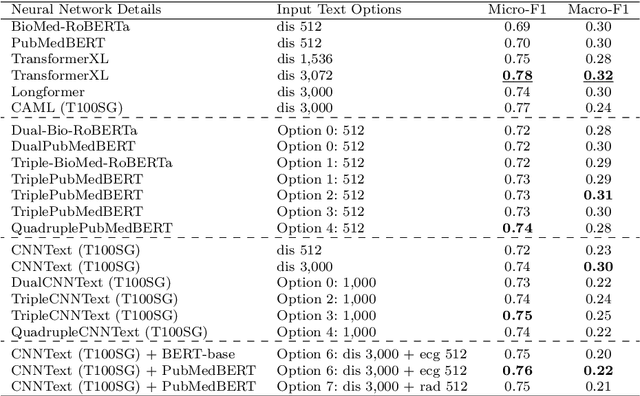
Abstract:Multi-label learning predicts a subset of labels from a given label set for an unseen instance while considering label correlations. A known challenge with multi-label classification is the long-tailed distribution of labels. Many studies focus on improving the overall predictions of the model and thus do not prioritise tail-end labels. Improving the tail-end label predictions in multi-label classifications of medical text enables the potential to understand patients better and improve care. The knowledge gained by one or more infrequent labels can impact the cause of medical decisions and treatment plans. This research presents variations of concatenated domain-specific language models, including multi-BioMed-Transformers, to achieve two primary goals. First, to improve F1 scores of infrequent labels across multi-label problems, especially with long-tail labels; second, to handle long medical text and multi-sourced electronic health records (EHRs), a challenging task for standard transformers designed to work on short input sequences. A vital contribution of this research is new state-of-the-art (SOTA) results obtained using TransformerXL for predicting medical codes. A variety of experiments are performed on the Medical Information Mart for Intensive Care (MIMIC-III) database. Results show that concatenated BioMed-Transformers outperform standard transformers in terms of overall micro and macro F1 scores and individual F1 scores of tail-end labels, while incurring lower training times than existing transformer-based solutions for long input sequences.
Predicting COVID-19 Patient Shielding: A Comprehensive Study
Oct 01, 2021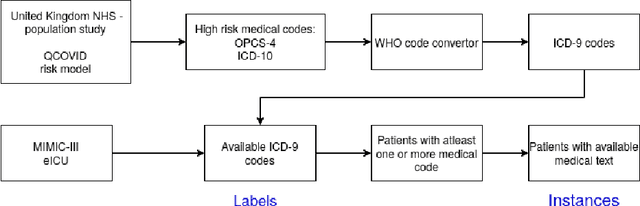
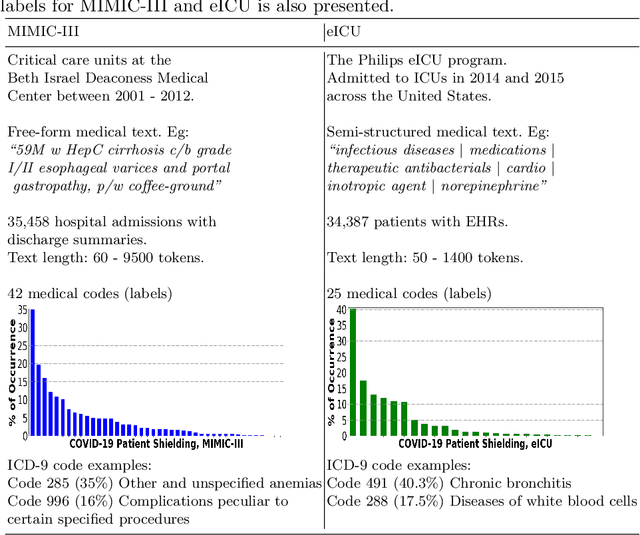
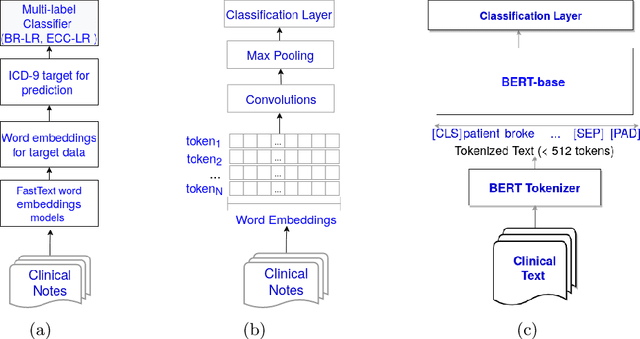
Abstract:There are many ways machine learning and big data analytics are used in the fight against the COVID-19 pandemic, including predictions, risk management, diagnostics, and prevention. This study focuses on predicting COVID-19 patient shielding -- identifying and protecting patients who are clinically extremely vulnerable from coronavirus. This study focuses on techniques used for the multi-label classification of medical text. Using the information published by the United Kingdom NHS and the World Health Organisation, we present a novel approach to predicting COVID-19 patient shielding as a multi-label classification problem. We use publicly available, de-identified ICU medical text data for our experiments. The labels are derived from the published COVID-19 patient shielding data. We present an extensive comparison across 12 multi-label classifiers from the simple binary relevance to neural networks and the most recent transformers. To the best of our knowledge this is the first comprehensive study, where such a range of multi-label classifiers for medical text are considered. We highlight the benefits of various approaches, and argue that, for the task at hand, both predictive accuracy and processing time are essential.
* Accepted in AJCAI 2021
Seeing The Whole Patient: Using Multi-Label Medical Text Classification Techniques to Enhance Predictions of Medical Codes
Mar 29, 2020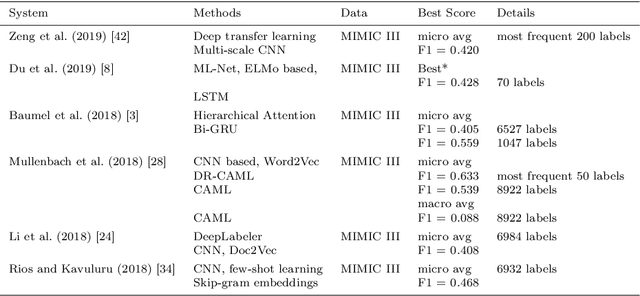
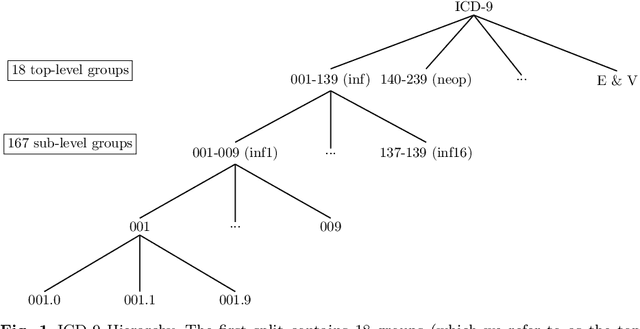
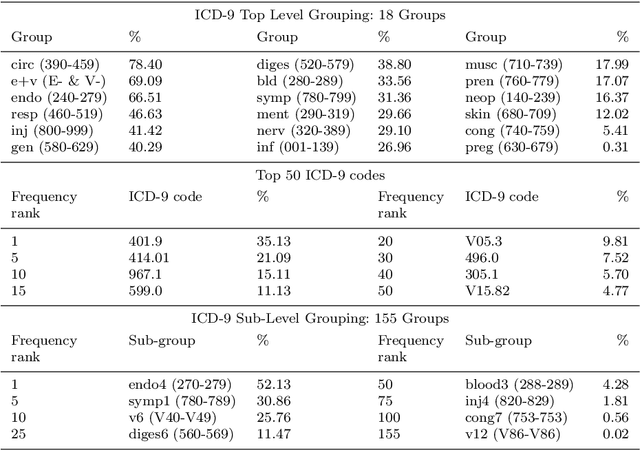
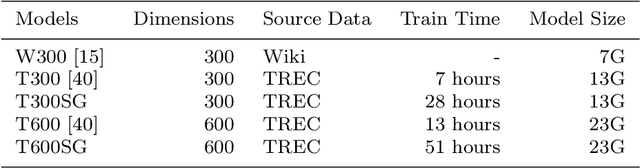
Abstract:Machine learning-based multi-label medical text classifications can be used to enhance the understanding of the human body and aid the need for patient care. We present a broad study on clinical natural language processing techniques to maximise a feature representing text when predicting medical codes on patients with multi-morbidity. We present results of multi-label medical text classification problems with 18, 50 and 155 labels. We compare several variations to embeddings, text tagging, and pre-processing. For imbalanced data we show that labels which occur infrequently, benefit the most from additional features incorporated in embeddings. We also show that high dimensional embeddings pre-trained using health-related data present a significant improvement in a multi-label setting, similarly to the way they improve performance for binary classification. High dimensional embeddings from this research are made available for public use.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge