Tingting Zheng
Spiking Meets Attention: Efficient Remote Sensing Image Super-Resolution with Attention Spiking Neural Networks
Mar 06, 2025Abstract:Spiking neural networks (SNNs) are emerging as a promising alternative to traditional artificial neural networks (ANNs), offering biological plausibility and energy efficiency. Despite these merits, SNNs are frequently hampered by limited capacity and insufficient representation power, yet remain underexplored in remote sensing super-resolution (SR) tasks. In this paper, we first observe that spiking signals exhibit drastic intensity variations across diverse textures, highlighting an active learning state of the neurons. This observation motivates us to apply SNNs for efficient SR of RSIs. Inspired by the success of attention mechanisms in representing salient information, we devise the spiking attention block (SAB), a concise yet effective component that optimizes membrane potentials through inferred attention weights, which, in turn, regulates spiking activity for superior feature representation. Our key contributions include: 1) we bridge the independent modulation between temporal and channel dimensions, facilitating joint feature correlation learning, and 2) we access the global self-similar patterns in large-scale remote sensing imagery to infer spatial attention weights, incorporating effective priors for realistic and faithful reconstruction. Building upon SAB, we proposed SpikeSR, which achieves state-of-the-art performance across various remote sensing benchmarks such as AID, DOTA, and DIOR, while maintaining high computational efficiency. The code of SpikeSR will be available upon paper acceptance.
Dynamic Policy-Driven Adaptive Multi-Instance Learning for Whole Slide Image Classification
Mar 09, 2024



Abstract:Multi-Instance Learning (MIL) has shown impressive performance for histopathology whole slide image (WSI) analysis using bags or pseudo-bags. It involves instance sampling, feature representation, and decision-making. However, existing MIL-based technologies at least suffer from one or more of the following problems: 1) requiring high storage and intensive pre-processing for numerous instances (sampling); 2) potential over-fitting with limited knowledge to predict bag labels (feature representation); 3) pseudo-bag counts and prior biases affect model robustness and generalizability (decision-making). Inspired by clinical diagnostics, using the past sampling instances can facilitate the final WSI analysis, but it is barely explored in prior technologies. To break free these limitations, we integrate the dynamic instance sampling and reinforcement learning into a unified framework to improve the instance selection and feature aggregation, forming a novel Dynamic Policy Instance Selection (DPIS) scheme for better and more credible decision-making. Specifically, the measurement of feature distance and reward function are employed to boost continuous instance sampling. To alleviate the over-fitting, we explore the latent global relations among instances for more robust and discriminative feature representation while establishing reward and punishment mechanisms to correct biases in pseudo-bags using contrastive learning. These strategies form the final Dynamic Policy-Driven Adaptive Multi-Instance Learning (PAMIL) method for WSI tasks. Extensive experiments reveal that our PAMIL method outperforms the state-of-the-art by 3.8\% on CAMELYON16 and 4.4\% on TCGA lung cancer datasets.
Global Contrast Masked Autoencoders Are Powerful Pathological Representation Learners
May 21, 2022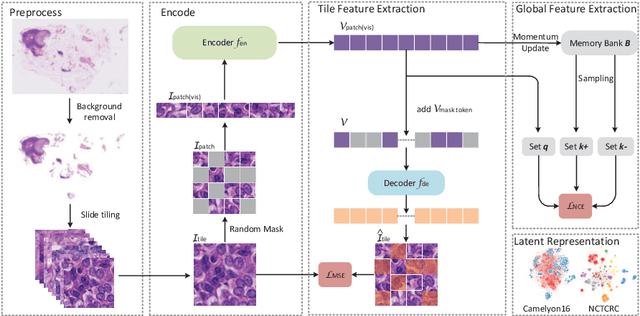
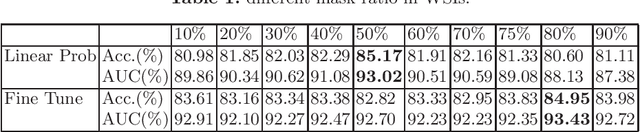
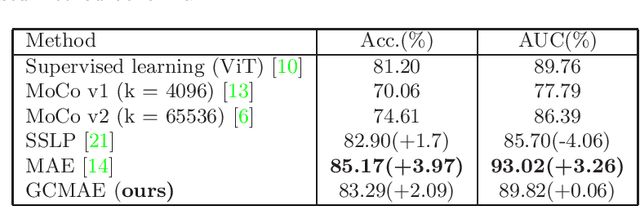
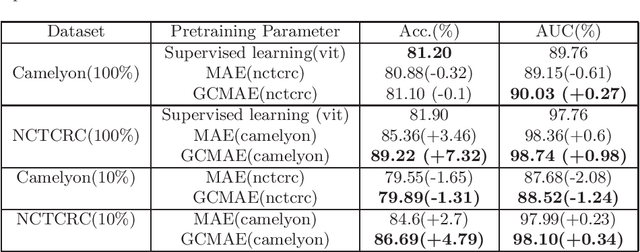
Abstract:Based on digital whole slide scanning technique, artificial intelligence algorithms represented by deep learning have achieved remarkable results in the field of computational pathology. Compared with other medical images such as Computed Tomography (CT) or Magnetic Resonance Imaging (MRI), pathological images are more difficult to annotate, thus there is an extreme lack of data sets that can be used for supervised learning. In this study, a self-supervised learning (SSL) model, Global Contrast Masked Autoencoders (GCMAE), is proposed, which has the ability to represent both global and local domain-specific features of whole slide image (WSI), as well as excellent cross-data transfer ability. The Camelyon16 and NCTCRC datasets are used to evaluate the performance of our model. When dealing with transfer learning tasks with different data sets, the experimental results show that GCMAE has better linear classification accuracy than MAE, which can reach 81.10% and 89.22% respectively. Our method outperforms the previous state-of-the-art algorithm and even surpass supervised learning (improved by 3.86% on NCTCRC data sets). The source code of this paper is publicly available at https://github.com/StarUniversus/gcmae
A Deep Reinforcement Learning Framework for Rapid Diagnosis of Whole Slide Pathological Images
May 05, 2022



Abstract:The deep neural network is a research hotspot for histopathological image analysis, which can improve the efficiency and accuracy of diagnosis for pathologists or be used for disease screening. The whole slide pathological image can reach one gigapixel and contains abundant tissue feature information, which needs to be divided into a lot of patches in the training and inference stages. This will lead to a long convergence time and large memory consumption. Furthermore, well-annotated data sets are also in short supply in the field of digital pathology. Inspired by the pathologist's clinical diagnosis process, we propose a weakly supervised deep reinforcement learning framework, which can greatly reduce the time required for network inference. We use neural network to construct the search model and decision model of reinforcement learning agent respectively. The search model predicts the next action through the image features of different magnifications in the current field of view, and the decision model is used to return the predicted probability of the current field of view image. In addition, an expert-guided model is constructed by multi-instance learning, which not only provides rewards for search model, but also guides decision model learning by the knowledge distillation method. Experimental results show that our proposed method can achieve fast inference and accurate prediction of whole slide images without any pixel-level annotations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge