Hao Quan
Distilling High Diagnostic Value Patches for Whole Slide Image Classification Using Attention Mechanism
Jul 29, 2024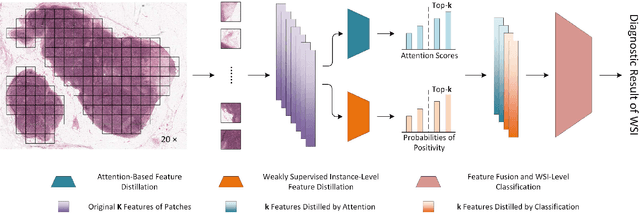
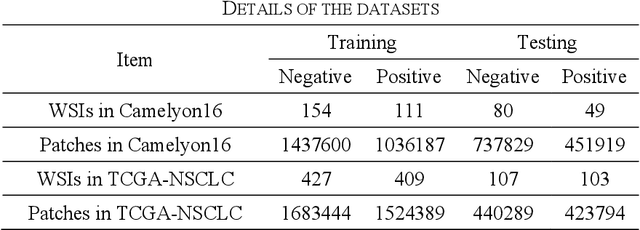
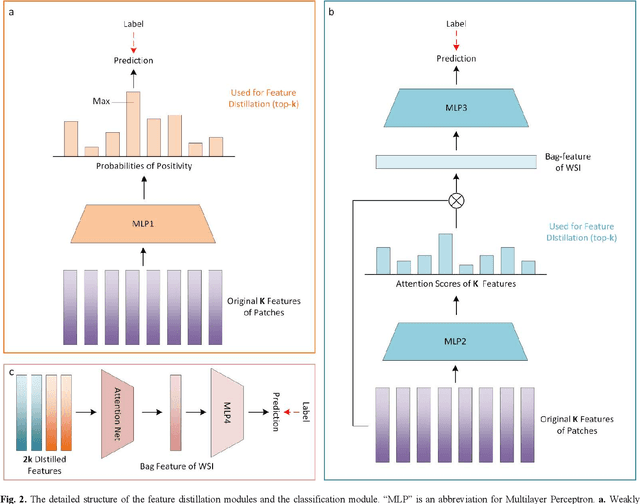
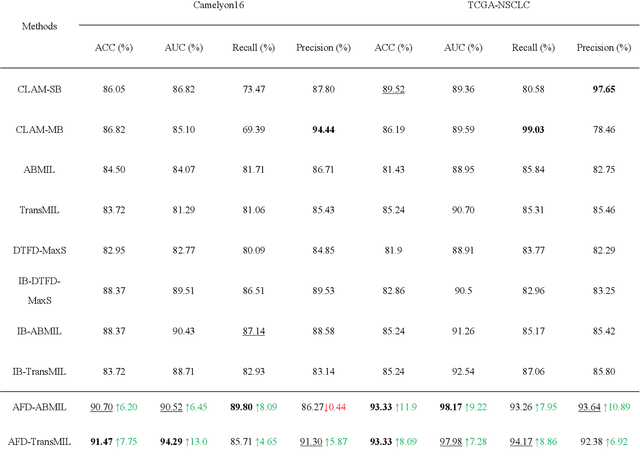
Abstract:Multiple Instance Learning (MIL) has garnered widespread attention in the field of Whole Slide Image (WSI) classification as it replaces pixel-level manual annotation with diagnostic reports as labels, significantly reducing labor costs. Recent research has shown that bag-level MIL methods often yield better results because they can consider all patches of the WSI as a whole. However, a drawback of such methods is the incorporation of more redundant patches, leading to interference. To extract patches with high diagnostic value while excluding interfering patches to address this issue, we developed an attention-based feature distillation multi-instance learning (AFD-MIL) approach. This approach proposed the exclusion of redundant patches as a preprocessing operation in weakly supervised learning, directly mitigating interference from extensive noise. It also pioneers the use of attention mechanisms to distill features with high diagnostic value, as opposed to the traditional practice of indiscriminately and forcibly integrating all patches. Additionally, we introduced global loss optimization to finely control the feature distillation module. AFD-MIL is orthogonal to many existing MIL methods, leading to consistent performance improvements. This approach has surpassed the current state-of-the-art method, achieving 91.47% ACC (accuracy) and 94.29% AUC (area under the curve) on the Camelyon16 (Camelyon Challenge 2016, breast cancer), while 93.33% ACC and 98.17% AUC on the TCGA-NSCLC (The Cancer Genome Atlas Program: non-small cell lung cancer). Different feature distillation methods were used for the two datasets, tailored to the specific diseases, thereby improving performance and interpretability.
Establishing Truly Causal Relationship Between Whole Slide Image Predictions and Diagnostic Evidence Subregions in Deep Learning
Jul 24, 2024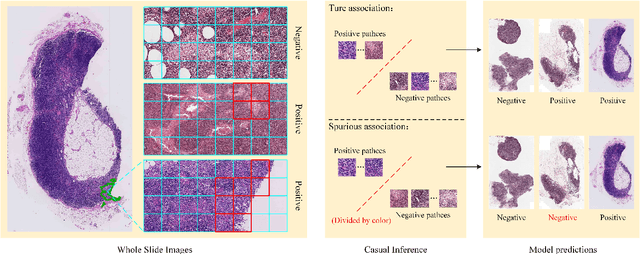
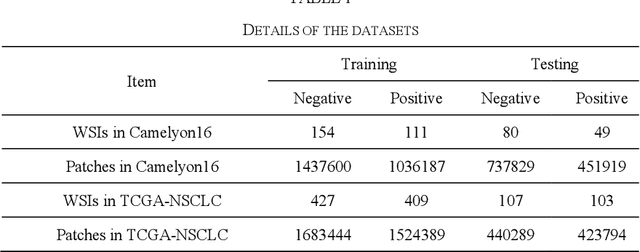

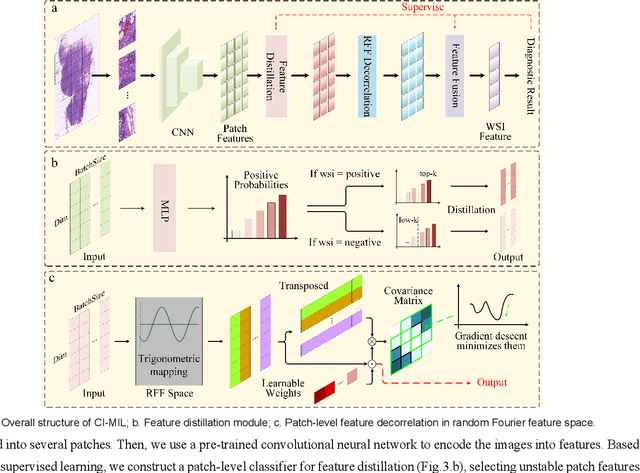
Abstract:In the field of deep learning-driven Whole Slide Image (WSI) classification, Multiple Instance Learning (MIL) has gained significant attention due to its ability to be trained using only slide-level diagnostic labels. Previous MIL researches have primarily focused on enhancing feature aggregators for globally analyzing WSIs, but overlook a causal relationship in diagnosis: model's prediction should ideally stem solely from regions of the image that contain diagnostic evidence (such as tumor cells), which usually occupy relatively small areas. To address this limitation and establish the truly causal relationship between model predictions and diagnostic evidence regions, we propose Causal Inference Multiple Instance Learning (CI-MIL). CI-MIL integrates feature distillation with a novel patch decorrelation mechanism, employing a two-stage causal inference approach to distill and process patches with high diagnostic value. Initially, CI-MIL leverages feature distillation to identify patches likely containing tumor cells and extracts their corresponding feature representations. These features are then mapped to random Fourier feature space, where a learnable weighting scheme is employed to minimize inter-feature correlations, effectively reducing redundancy from homogenous patches and mitigating data bias. These processes strengthen the causal relationship between model predictions and diagnostically relevant regions, making the prediction more direct and reliable. Experimental results demonstrate that CI-MIL outperforms state-of-the-art methods. Additionally, CI-MIL exhibits superior interpretability, as its selected regions demonstrate high consistency with ground truth annotations, promising more reliable diagnostic assistance for pathologists.
Dual-channel Prototype Network for few-shot Classification of Pathological Images
Nov 14, 2023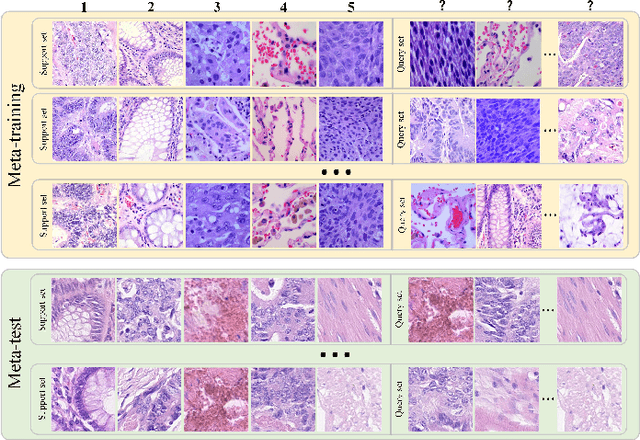
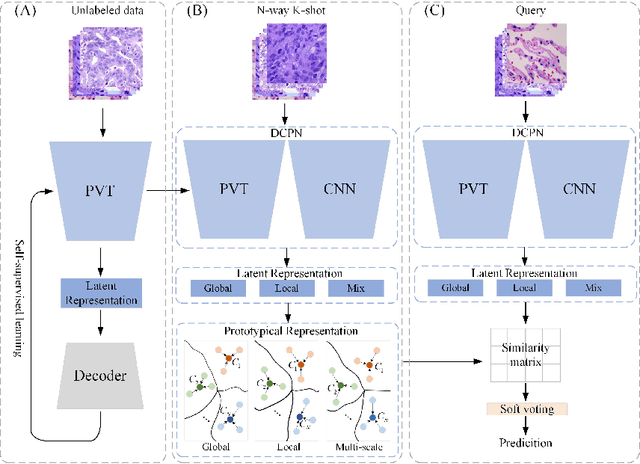
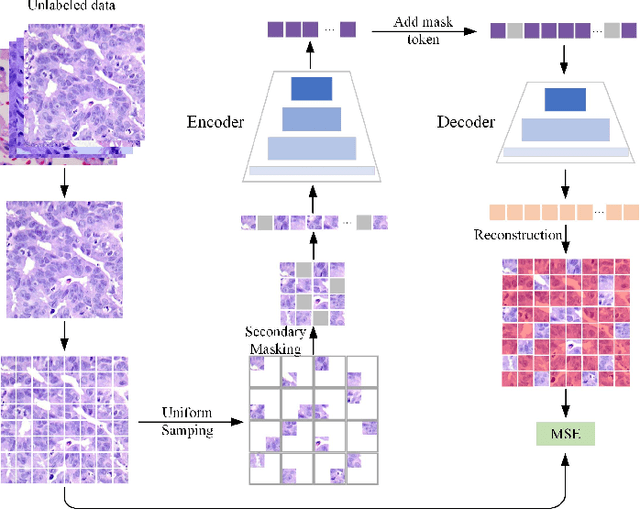
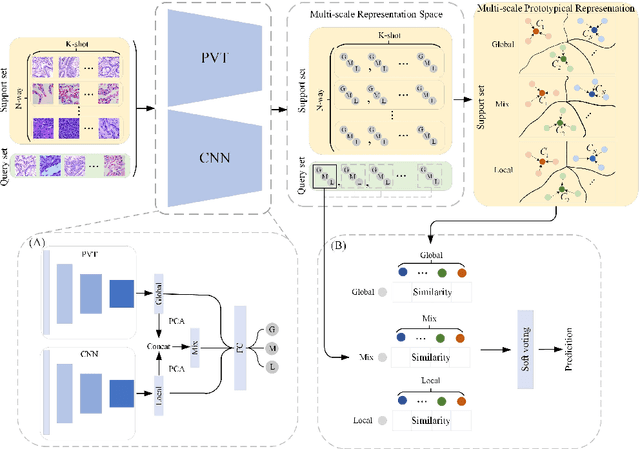
Abstract:In pathology, the rarity of certain diseases and the complexity in annotating pathological images significantly hinder the creation of extensive, high-quality datasets. This limitation impedes the progress of deep learning-assisted diagnostic systems in pathology. Consequently, it becomes imperative to devise a technology that can discern new disease categories from a minimal number of annotated examples. Such a technology would substantially advance deep learning models for rare diseases. Addressing this need, we introduce the Dual-channel Prototype Network (DCPN), rooted in the few-shot learning paradigm, to tackle the challenge of classifying pathological images with limited samples. DCPN augments the Pyramid Vision Transformer (PVT) framework for few-shot classification via self-supervised learning and integrates it with convolutional neural networks. This combination forms a dual-channel architecture that extracts multi-scale, highly precise pathological features. The approach enhances the versatility of prototype representations and elevates the efficacy of prototype networks in few-shot pathological image classification tasks. We evaluated DCPN using three publicly available pathological datasets, configuring small-sample classification tasks that mirror varying degrees of clinical scenario domain shifts. Our experimental findings robustly affirm DCPN's superiority in few-shot pathological image classification, particularly in tasks within the same domain, where it achieves the benchmarks of supervised learning.
Global Contrast Masked Autoencoders Are Powerful Pathological Representation Learners
May 21, 2022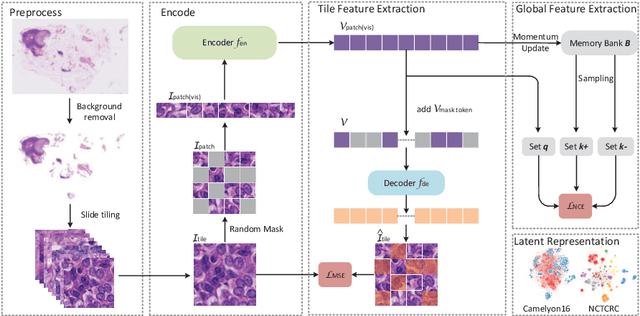
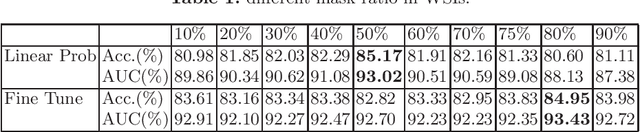
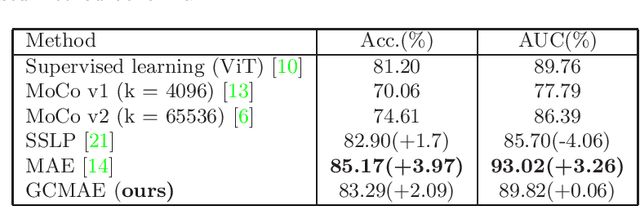
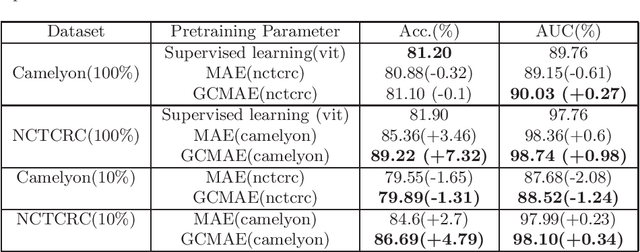
Abstract:Based on digital whole slide scanning technique, artificial intelligence algorithms represented by deep learning have achieved remarkable results in the field of computational pathology. Compared with other medical images such as Computed Tomography (CT) or Magnetic Resonance Imaging (MRI), pathological images are more difficult to annotate, thus there is an extreme lack of data sets that can be used for supervised learning. In this study, a self-supervised learning (SSL) model, Global Contrast Masked Autoencoders (GCMAE), is proposed, which has the ability to represent both global and local domain-specific features of whole slide image (WSI), as well as excellent cross-data transfer ability. The Camelyon16 and NCTCRC datasets are used to evaluate the performance of our model. When dealing with transfer learning tasks with different data sets, the experimental results show that GCMAE has better linear classification accuracy than MAE, which can reach 81.10% and 89.22% respectively. Our method outperforms the previous state-of-the-art algorithm and even surpass supervised learning (improved by 3.86% on NCTCRC data sets). The source code of this paper is publicly available at https://github.com/StarUniversus/gcmae
A Deep Reinforcement Learning Framework for Rapid Diagnosis of Whole Slide Pathological Images
May 05, 2022



Abstract:The deep neural network is a research hotspot for histopathological image analysis, which can improve the efficiency and accuracy of diagnosis for pathologists or be used for disease screening. The whole slide pathological image can reach one gigapixel and contains abundant tissue feature information, which needs to be divided into a lot of patches in the training and inference stages. This will lead to a long convergence time and large memory consumption. Furthermore, well-annotated data sets are also in short supply in the field of digital pathology. Inspired by the pathologist's clinical diagnosis process, we propose a weakly supervised deep reinforcement learning framework, which can greatly reduce the time required for network inference. We use neural network to construct the search model and decision model of reinforcement learning agent respectively. The search model predicts the next action through the image features of different magnifications in the current field of view, and the decision model is used to return the predicted probability of the current field of view image. In addition, an expert-guided model is constructed by multi-instance learning, which not only provides rewards for search model, but also guides decision model learning by the knowledge distillation method. Experimental results show that our proposed method can achieve fast inference and accurate prediction of whole slide images without any pixel-level annotations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge