Distilling High Diagnostic Value Patches for Whole Slide Image Classification Using Attention Mechanism
Paper and Code
Jul 29, 2024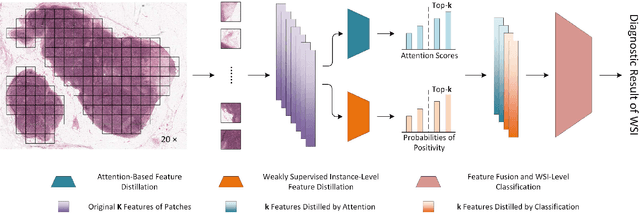
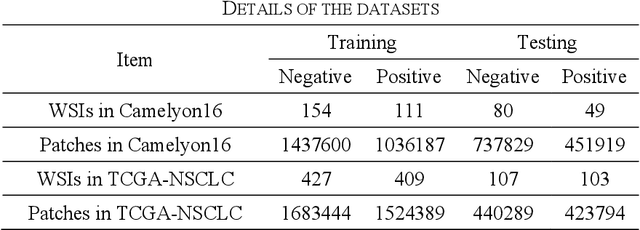
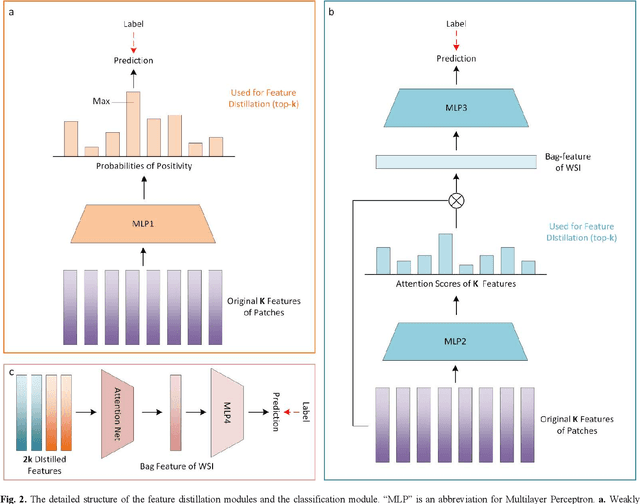
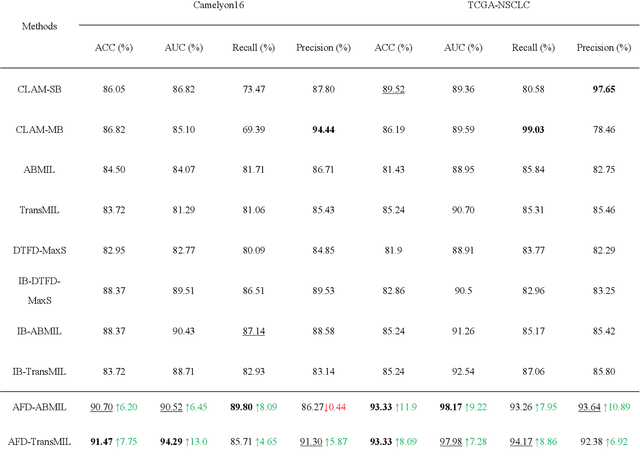
Multiple Instance Learning (MIL) has garnered widespread attention in the field of Whole Slide Image (WSI) classification as it replaces pixel-level manual annotation with diagnostic reports as labels, significantly reducing labor costs. Recent research has shown that bag-level MIL methods often yield better results because they can consider all patches of the WSI as a whole. However, a drawback of such methods is the incorporation of more redundant patches, leading to interference. To extract patches with high diagnostic value while excluding interfering patches to address this issue, we developed an attention-based feature distillation multi-instance learning (AFD-MIL) approach. This approach proposed the exclusion of redundant patches as a preprocessing operation in weakly supervised learning, directly mitigating interference from extensive noise. It also pioneers the use of attention mechanisms to distill features with high diagnostic value, as opposed to the traditional practice of indiscriminately and forcibly integrating all patches. Additionally, we introduced global loss optimization to finely control the feature distillation module. AFD-MIL is orthogonal to many existing MIL methods, leading to consistent performance improvements. This approach has surpassed the current state-of-the-art method, achieving 91.47% ACC (accuracy) and 94.29% AUC (area under the curve) on the Camelyon16 (Camelyon Challenge 2016, breast cancer), while 93.33% ACC and 98.17% AUC on the TCGA-NSCLC (The Cancer Genome Atlas Program: non-small cell lung cancer). Different feature distillation methods were used for the two datasets, tailored to the specific diseases, thereby improving performance and interpretability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge