Global Contrast Masked Autoencoders Are Powerful Pathological Representation Learners
Paper and Code
May 21, 2022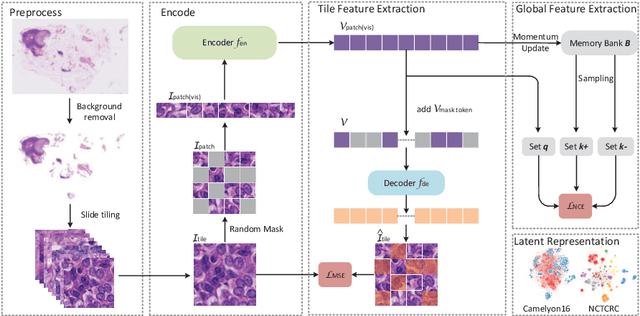
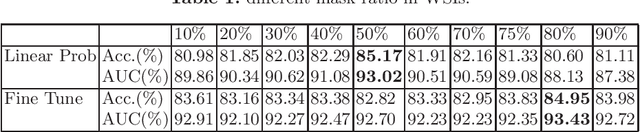
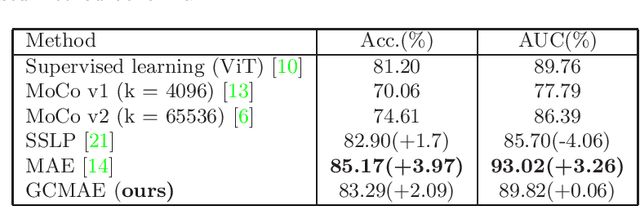
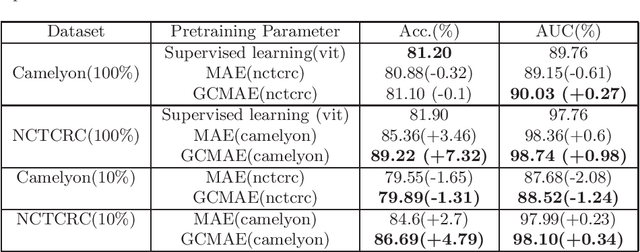
Based on digital whole slide scanning technique, artificial intelligence algorithms represented by deep learning have achieved remarkable results in the field of computational pathology. Compared with other medical images such as Computed Tomography (CT) or Magnetic Resonance Imaging (MRI), pathological images are more difficult to annotate, thus there is an extreme lack of data sets that can be used for supervised learning. In this study, a self-supervised learning (SSL) model, Global Contrast Masked Autoencoders (GCMAE), is proposed, which has the ability to represent both global and local domain-specific features of whole slide image (WSI), as well as excellent cross-data transfer ability. The Camelyon16 and NCTCRC datasets are used to evaluate the performance of our model. When dealing with transfer learning tasks with different data sets, the experimental results show that GCMAE has better linear classification accuracy than MAE, which can reach 81.10% and 89.22% respectively. Our method outperforms the previous state-of-the-art algorithm and even surpass supervised learning (improved by 3.86% on NCTCRC data sets). The source code of this paper is publicly available at https://github.com/StarUniversus/gcmae
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge