Tiberiu Tesileanu
xVal: A Continuous Number Encoding for Large Language Models
Oct 04, 2023Abstract:Large Language Models have not yet been broadly adapted for the analysis of scientific datasets due in part to the unique difficulties of tokenizing numbers. We propose xVal, a numerical encoding scheme that represents any real number using just a single token. xVal represents a given real number by scaling a dedicated embedding vector by the number value. Combined with a modified number-inference approach, this strategy renders the model end-to-end continuous when considered as a map from the numbers of the input string to those of the output string. This leads to an inductive bias that is generally more suitable for applications in scientific domains. We empirically evaluate our proposal on a number of synthetic and real-world datasets. Compared with existing number encoding schemes, we find that xVal is more token-efficient and demonstrates improved generalization.
Multiple Physics Pretraining for Physical Surrogate Models
Oct 04, 2023


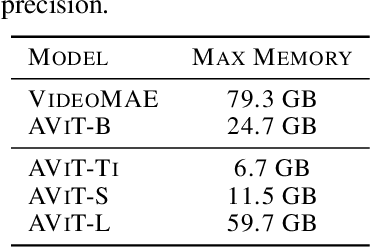
Abstract:We introduce multiple physics pretraining (MPP), an autoregressive task-agnostic pretraining approach for physical surrogate modeling. MPP involves training large surrogate models to predict the dynamics of multiple heterogeneous physical systems simultaneously by learning features that are broadly useful across diverse physical tasks. In order to learn effectively in this setting, we introduce a shared embedding and normalization strategy that projects the fields of multiple systems into a single shared embedding space. We validate the efficacy of our approach on both pretraining and downstream tasks over a broad fluid mechanics-oriented benchmark. We show that a single MPP-pretrained transformer is able to match or outperform task-specific baselines on all pretraining sub-tasks without the need for finetuning. For downstream tasks, we demonstrate that finetuning MPP-trained models results in more accurate predictions across multiple time-steps on new physics compared to training from scratch or finetuning pretrained video foundation models. We open-source our code and model weights trained at multiple scales for reproducibility and community experimentation.
AstroCLIP: Cross-Modal Pre-Training for Astronomical Foundation Models
Oct 04, 2023Abstract:We present AstroCLIP, a strategy to facilitate the construction of astronomical foundation models that bridge the gap between diverse observational modalities. We demonstrate that a cross-modal contrastive learning approach between images and optical spectra of galaxies yields highly informative embeddings of both modalities. In particular, we apply our method on multi-band images and optical spectra from the Dark Energy Spectroscopic Instrument (DESI), and show that: (1) these embeddings are well-aligned between modalities and can be used for accurate cross-modal searches, and (2) these embeddings encode valuable physical information about the galaxies -- in particular redshift and stellar mass -- that can be used to achieve competitive zero- and few- shot predictions without further finetuning. Additionally, in the process of developing our approach, we also construct a novel, transformer-based model and pretraining approach for processing galaxy spectra.
Reusability report: Prostate cancer stratification with diverse biologically-informed neural architectures
Sep 28, 2023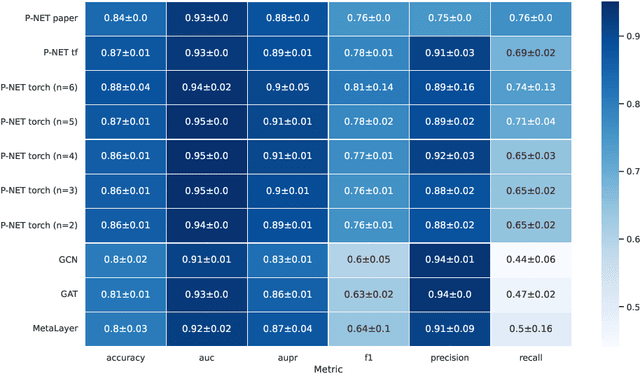
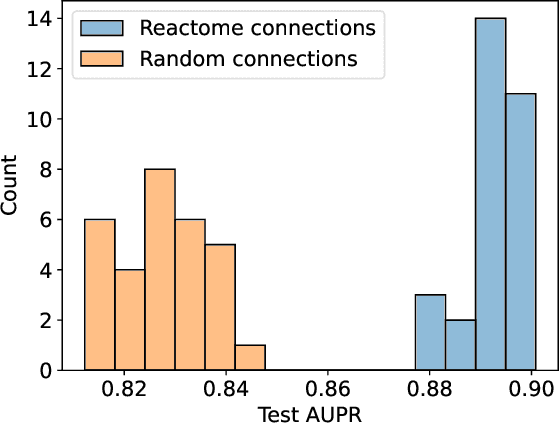
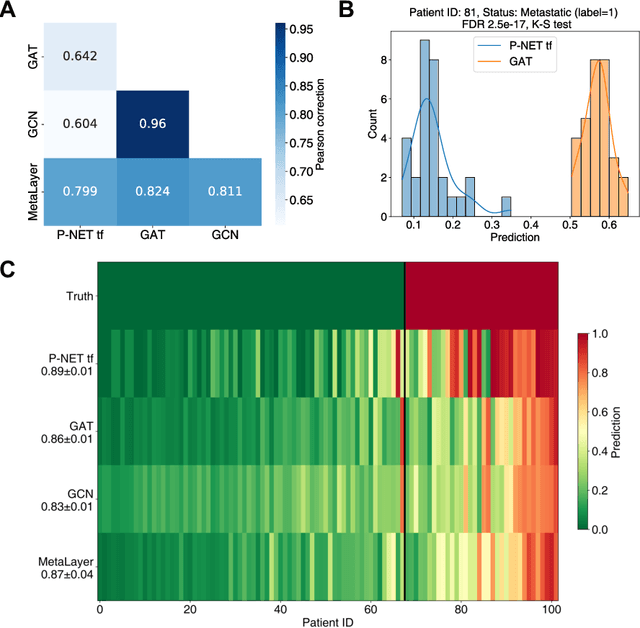
Abstract:In, Elmarakeby et al., "Biologically informed deep neural network for prostate cancer discovery", a feedforward neural network with biologically informed, sparse connections (P-NET) was presented to model the state of prostate cancer. We verified the reproducibility of the study conducted by Elmarakeby et al., using both their original codebase, and our own re-implementation using more up-to-date libraries. We quantified the contribution of network sparsification by Reactome biological pathways, and confirmed its importance to P-NET's superior performance. Furthermore, we explored alternative neural architectures and approaches to incorporating biological information into the networks. We experimented with three types of graph neural networks on the same training data, and investigated the clinical prediction agreement between different models. Our analyses demonstrated that deep neural networks with distinct architectures make incorrect predictions for individual patient that are persistent across different initializations of a specific neural architecture. This suggests that different neural architectures are sensitive to different aspects of the data, an important yet under-explored challenge for clinical prediction tasks.
An online algorithm for contrastive Principal Component Analysis
Nov 14, 2022



Abstract:Finding informative low-dimensional representations that can be computed efficiently in large datasets is an important problem in data analysis. Recently, contrastive Principal Component Analysis (cPCA) was proposed as a more informative generalization of PCA that takes advantage of contrastive learning. However, the performance of cPCA is sensitive to hyper-parameter choice and there is currently no online algorithm for implementing cPCA. Here, we introduce a modified cPCA method, which we denote cPCA*, that is more interpretable and less sensitive to the choice of hyper-parameter. We derive an online algorithm for cPCA* and show that it maps onto a neural network with local learning rules, so it can potentially be implemented in energy efficient neuromorphic hardware. We evaluate the performance of our online algorithm on real datasets and highlight the differences and similarities with the original formulation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge