Reusability report: Prostate cancer stratification with diverse biologically-informed neural architectures
Paper and Code
Sep 28, 2023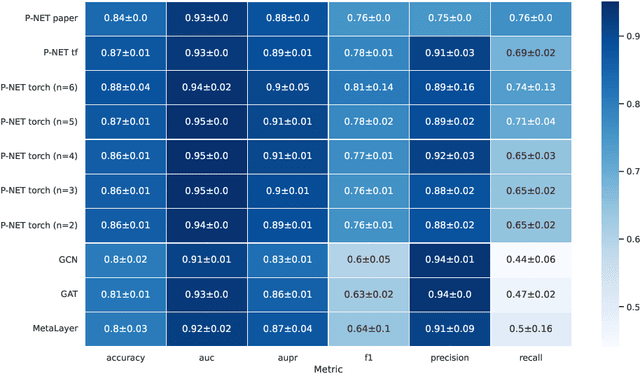
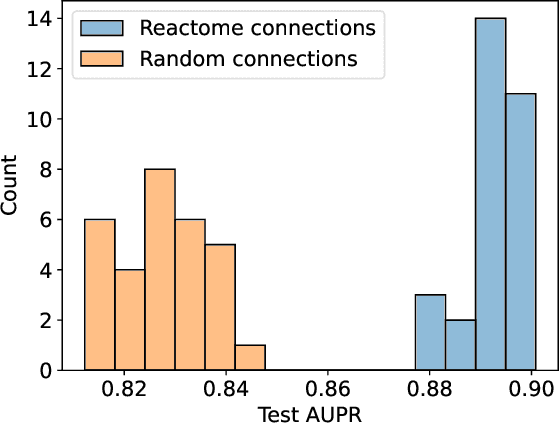
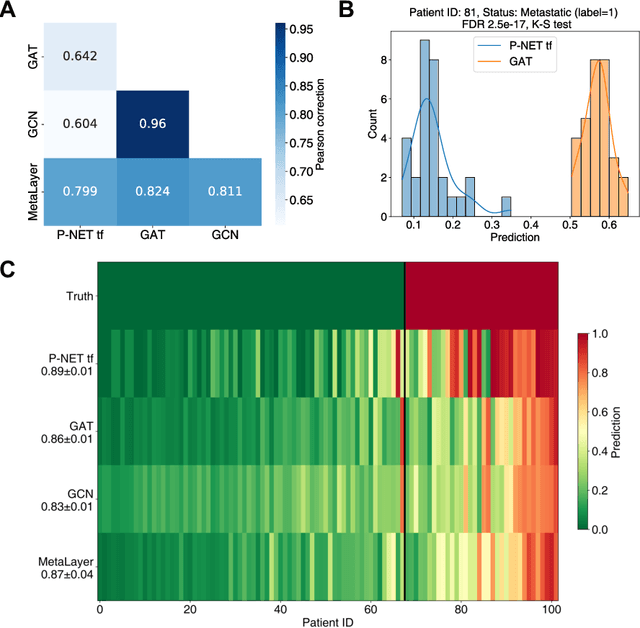
In, Elmarakeby et al., "Biologically informed deep neural network for prostate cancer discovery", a feedforward neural network with biologically informed, sparse connections (P-NET) was presented to model the state of prostate cancer. We verified the reproducibility of the study conducted by Elmarakeby et al., using both their original codebase, and our own re-implementation using more up-to-date libraries. We quantified the contribution of network sparsification by Reactome biological pathways, and confirmed its importance to P-NET's superior performance. Furthermore, we explored alternative neural architectures and approaches to incorporating biological information into the networks. We experimented with three types of graph neural networks on the same training data, and investigated the clinical prediction agreement between different models. Our analyses demonstrated that deep neural networks with distinct architectures make incorrect predictions for individual patient that are persistent across different initializations of a specific neural architecture. This suggests that different neural architectures are sensitive to different aspects of the data, an important yet under-explored challenge for clinical prediction tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge