Suvidha Tripathi
FRCSyn Challenge at WACV 2024:Face Recognition Challenge in the Era of Synthetic Data
Nov 17, 2023



Abstract:Despite the widespread adoption of face recognition technology around the world, and its remarkable performance on current benchmarks, there are still several challenges that must be covered in more detail. This paper offers an overview of the Face Recognition Challenge in the Era of Synthetic Data (FRCSyn) organized at WACV 2024. This is the first international challenge aiming to explore the use of synthetic data in face recognition to address existing limitations in the technology. Specifically, the FRCSyn Challenge targets concerns related to data privacy issues, demographic biases, generalization to unseen scenarios, and performance limitations in challenging scenarios, including significant age disparities between enrollment and testing, pose variations, and occlusions. The results achieved in the FRCSyn Challenge, together with the proposed benchmark, contribute significantly to the application of synthetic data to improve face recognition technology.
MUNCH: Modelling Unique 'N Controllable Heads
Oct 04, 2023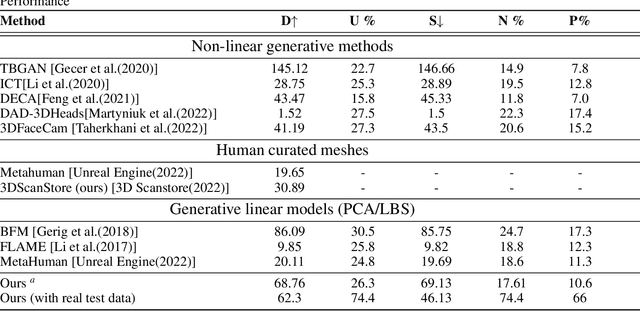
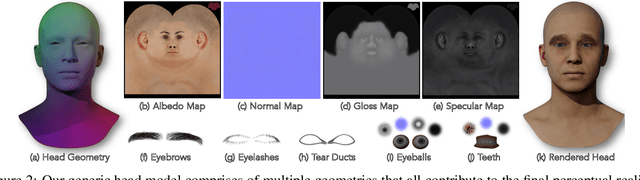
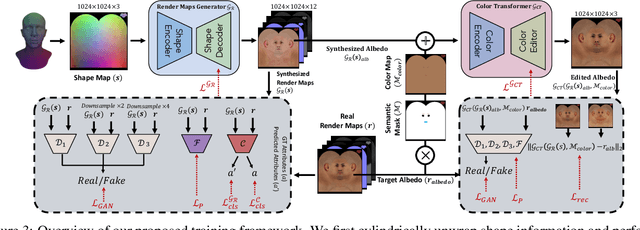

Abstract:The automated generation of 3D human heads has been an intriguing and challenging task for computer vision researchers. Prevailing methods synthesize realistic avatars but with limited control over the diversity and quality of rendered outputs and suffer from limited correlation between shape and texture of the character. We propose a method that offers quality, diversity, control, and realism along with explainable network design, all desirable features to game-design artists in the domain. First, our proposed Geometry Generator identifies disentangled latent directions and generate novel and diverse samples. A Render Map Generator then learns to synthesize multiply high-fidelty physically-based render maps including Albedo, Glossiness, Specular, and Normals. For artists preferring fine-grained control over the output, we introduce a novel Color Transformer Model that allows semantic color control over generated maps. We also introduce quantifiable metrics called Uniqueness and Novelty and a combined metric to test the overall performance of our model. Demo for both shapes and textures can be found: https://munch-seven.vercel.app/. We will release our model along with the synthetic dataset.
Bag of Visual Words with Deep Features -- Patch Classification Model for Limited Dataset of Breast Tumours
Feb 22, 2022
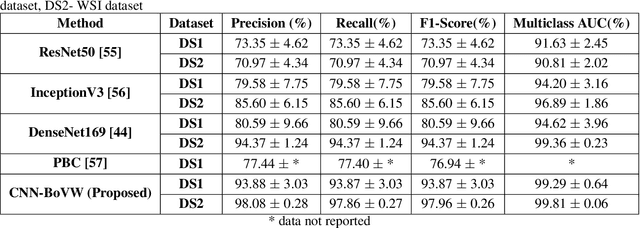
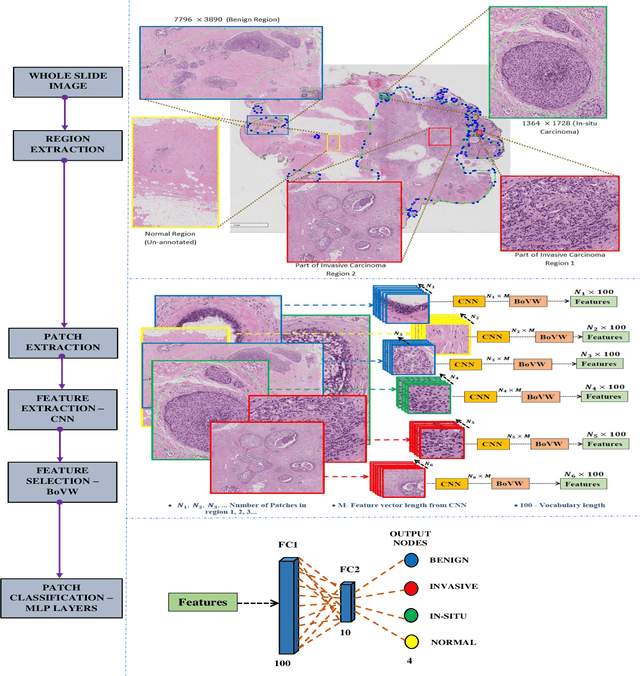
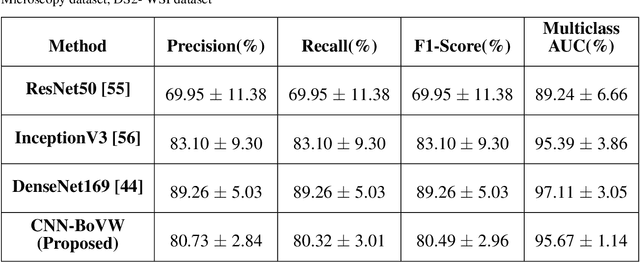
Abstract:Currently, the computational complexity limits the training of high resolution gigapixel images using Convolutional Neural Networks. Therefore, such images are divided into patches or tiles. Since, these high resolution patches are encoded with discriminative information therefore; CNNs are trained on these patches to perform patch-level predictions. However, the problem with patch-level prediction is that pathologist generally annotates at image-level and not at patch level. Due to this limitation most of the patches may not contain enough class-relevant features. Through this work, we tried to incorporate patch descriptive capability within the deep framework by using Bag of Visual Words (BoVW) as a kind of regularisation to improve generalizability. Using this hypothesis, we aim to build a patch based classifier to discriminate between four classes of breast biopsy image patches (normal, benign, \textit{In situ} carcinoma, invasive carcinoma). The task is to incorporate quality deep features using CNN to describe relevant information in the images while simultaneously discarding irrelevant information using Bag of Visual Words (BoVW). The proposed method passes patches obtained from WSI and microscopy images through pre-trained CNN to extract features. BoVW is used as a feature selector to select most discriminative features among the CNN features. Finally, the selected feature sets are classified as one of the four classes. The hybrid model provides flexibility in terms of choice of pre-trained models for feature extraction. The pipeline is end-to-end since it does not require post processing of patch predictions to select discriminative patches. We compared our observations with state-of-the-art methods like ResNet50, DenseNet169, and InceptionV3 on the BACH-2018 challenge dataset. Our proposed method shows better performance than all the three methods.
Ensembling Handcrafted Features with Deep Features: An Analytical Study for Classification of Routine Colon Cancer Histopathological Nuclei Images
Feb 22, 2022

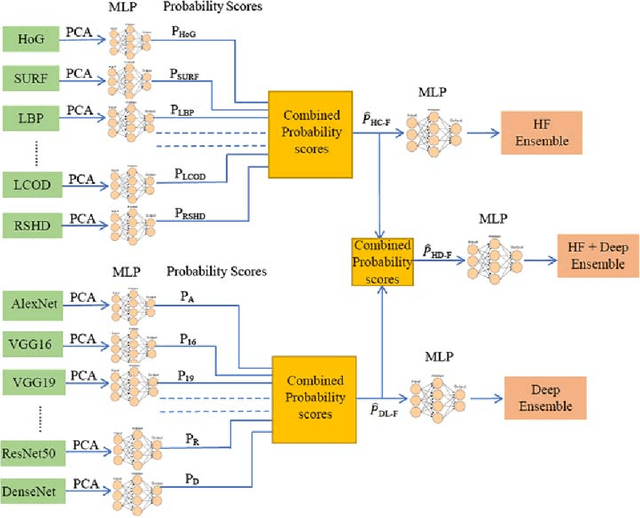

Abstract:The use of Deep Learning (DL) based methods in medical histopathology images have been one of the most sought after solutions to classify, segment, and detect diseased biopsy samples. However, given the complex nature of medical datasets due to the presence of intra-class variability and heterogeneity, the use of complex DL models might not give the optimal performance up to the level which is suitable for assisting pathologists. Therefore, ensemble DL methods with the scope of including domain agnostic handcrafted Features (HC-F) inspired this work. We have, through experiments, tried to highlight that a single DL network (domain-specific or state of the art pre-trained models) cannot be directly used as the base model without proper analysis with the relevant dataset. We have used F1-measure, Precision, Recall, AUC, and Cross-Entropy Loss to analyse the performance of our approaches. We observed from the results that the DL features ensemble bring a marked improvement in the overall performance of the model, whereas, domain agnostic HC-F remains dormant on the performance of the DL models.
An Object Aware Hybrid U-Net for Breast Tumour Annotation
Feb 22, 2022Abstract:In the clinical settings, during digital examination of histopathological slides, the pathologist annotate the slides by marking the rough boundary around the suspected tumour region. The marking or annotation is generally represented as a polygonal boundary that covers the extent of the tumour in the slide. These polygonal markings are difficult to imitate through CAD techniques since the tumour regions are heterogeneous and hence segmenting them would require exhaustive pixel wise ground truth annotation. Therefore, for CAD analysis, the ground truths are generally annotated by pathologist explicitly for research purposes. However, this kind of annotation which is generally required for semantic or instance segmentation is time consuming and tedious. In this proposed work, therefore, we have tried to imitate pathologist like annotation by segmenting tumour extents by polygonal boundaries. For polygon like annotation or segmentation, we have used Active Contours whose vertices or snake points move towards the boundary of the object of interest to find the region of minimum energy. To penalize the Active Contour we used modified U-Net architecture for learning penalization values. The proposed hybrid deep learning model fuses the modern deep learning segmentation algorithm with traditional Active Contours segmentation technique. The model is tested against both state-of-the-art semantic segmentation and hybrid models for performance evaluation against contemporary work. The results obtained show that the pathologist like annotation could be achieved by developing such hybrid models that integrate the domain knowledge through classical segmentation methods like Active Contours and global knowledge through semantic segmentation deep learning models.
Cell nuclei classification in histopathological images using hybrid OLConvNet
Feb 21, 2022



Abstract:Computer-aided histopathological image analysis for cancer detection is a major research challenge in the medical domain. Automatic detection and classification of nuclei for cancer diagnosis impose a lot of challenges in developing state of the art algorithms due to the heterogeneity of cell nuclei and data set variability. Recently, a multitude of classification algorithms has used complex deep learning models for their dataset. However, most of these methods are rigid and their architectural arrangement suffers from inflexibility and non-interpretability. In this research article, we have proposed a hybrid and flexible deep learning architecture OLConvNet that integrates the interpretability of traditional object-level features and generalization of deep learning features by using a shallower Convolutional Neural Network (CNN) named as $CNN_{3L}$. $CNN_{3L}$ reduces the training time by training fewer parameters and hence eliminating space constraints imposed by deeper algorithms. We used F1-score and multiclass Area Under the Curve (AUC) performance parameters to compare the results. To further strengthen the viability of our architectural approach, we tested our proposed methodology with state of the art deep learning architectures AlexNet, VGG16, VGG19, ResNet50, InceptionV3, and DenseNet121 as backbone networks. After a comprehensive analysis of classification results from all four architectures, we observed that our proposed model works well and perform better than contemporary complex algorithms.
An End-to-End Breast Tumour Classification Model Using Context-Based Patch Modelling- A BiLSTM Approach for Image Classification
Jun 05, 2021



Abstract:Researchers working on computational analysis of Whole Slide Images (WSIs) in histopathology have primarily resorted to patch-based modelling due to large resolution of each WSI. The large resolution makes WSIs infeasible to be fed directly into the machine learning models due to computational constraints. However, due to patch-based analysis, most of the current methods fail to exploit the underlying spatial relationship among the patches. In our work, we have tried to integrate this relationship along with feature-based correlation among the extracted patches from the particular tumorous region. For the given task of classification, we have used BiLSTMs to model both forward and backward contextual relationship. RNN based models eliminate the limitation of sequence size by allowing the modelling of variable size images within a deep learning model. We have also incorporated the effect of spatial continuity by exploring different scanning techniques used to sample patches. To establish the efficiency of our approach, we trained and tested our model on two datasets, microscopy images and WSI tumour regions. After comparing with contemporary literature we achieved the better performance with accuracy of 90% for microscopy image dataset. For WSI tumour region dataset, we compared the classification results with deep learning networks such as ResNet, DenseNet, and InceptionV3 using maximum voting technique. We achieved the highest performance accuracy of 84%. We found out that BiLSTMs with CNN features have performed much better in modelling patches into an end-to-end Image classification network. Additionally, the variable dimensions of WSI tumour regions were used for classification without the need for resizing. This suggests that our method is independent of tumour image size and can process large dimensional images without losing the resolution details.
* 36 pages, 5 figures, 9 tables. Published in Computerized Medical Imaging and Graphics
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge