Sunho Kim
Back to the Future: Look-ahead Augmentation and Parallel Self-Refinement for Time Series Forecasting
Feb 02, 2026Abstract:Long-term time series forecasting (LTSF) remains challenging due to the trade-off between parallel efficiency and sequential modeling of temporal coherence. Direct multi-step forecasting (DMS) methods enable fast, parallel prediction of all future horizons but often lose temporal consistency across steps, while iterative multi-step forecasting (IMS) preserves temporal dependencies at the cost of error accumulation and slow inference. To bridge this gap, we propose Back to the Future (BTTF), a simple yet effective framework that enhances forecasting stability through look-ahead augmentation and self-corrective refinement. Rather than relying on complex model architectures, BTTF revisits the fundamental forecasting process and refines a base model by ensembling the second-stage models augmented with their initial predictions. Despite its simplicity, our approach consistently improves long-horizon accuracy and mitigates the instability of linear forecasting models, achieving accuracy gains of up to 58% and demonstrating stable improvements even when the first-stage model is trained under suboptimal conditions. These results suggest that leveraging model-generated forecasts as augmentation can be a simple yet powerful way to enhance long-term prediction, even without complex architectures.
CREAM: Continual Retrieval on Dynamic Streaming Corpora with Adaptive Soft Memory
Jan 06, 2026Abstract:Information retrieval (IR) in dynamic data streams is emerging as a challenging task, as shifts in data distribution degrade the performance of AI-powered IR systems. To mitigate this issue, memory-based continual learning has been widely adopted for IR. However, existing methods rely on a fixed set of queries with ground-truth relevant documents, which limits generalization to unseen queries and documents, making them impractical for real-world applications. To enable more effective learning with unseen topics of a new corpus without ground-truth labels, we propose CREAM, a self-supervised framework for memory-based continual retrieval. CREAM captures the evolving semantics of streaming queries and documents into dynamically structured soft memory and leverages it to adapt to both seen and unseen topics in an unsupervised setting. We realize this through three key techniques: fine-grained similarity estimation, regularized cluster prototyping, and stratified coreset sampling. Experiments on two benchmark datasets demonstrate that CREAM exhibits superior adaptability and retrieval accuracy, outperforming the strongest method in a label-free setting by 27.79\% in Success@5 and 44.5\% in Recall@10 on average, and achieving performance comparable to or even exceeding that of supervised methods.
Enhancing Rating-Based Reinforcement Learning to Effectively Leverage Feedback from Large Vision-Language Models
Jun 15, 2025



Abstract:Designing effective reward functions remains a fundamental challenge in reinforcement learning (RL), as it often requires extensive human effort and domain expertise. While RL from human feedback has been successful in aligning agents with human intent, acquiring high-quality feedback is costly and labor-intensive, limiting its scalability. Recent advancements in foundation models present a promising alternative--leveraging AI-generated feedback to reduce reliance on human supervision in reward learning. Building on this paradigm, we introduce ERL-VLM, an enhanced rating-based RL method that effectively learns reward functions from AI feedback. Unlike prior methods that rely on pairwise comparisons, ERL-VLM queries large vision-language models (VLMs) for absolute ratings of individual trajectories, enabling more expressive feedback and improved sample efficiency. Additionally, we propose key enhancements to rating-based RL, addressing instability issues caused by data imbalance and noisy labels. Through extensive experiments across both low-level and high-level control tasks, we demonstrate that ERL-VLM significantly outperforms existing VLM-based reward generation methods. Our results demonstrate the potential of AI feedback for scaling RL with minimal human intervention, paving the way for more autonomous and efficient reward learning.
Bag of Tricks for Developing Diabetic Retinopathy Analysis Framework to Overcome Data Scarcity
Oct 18, 2022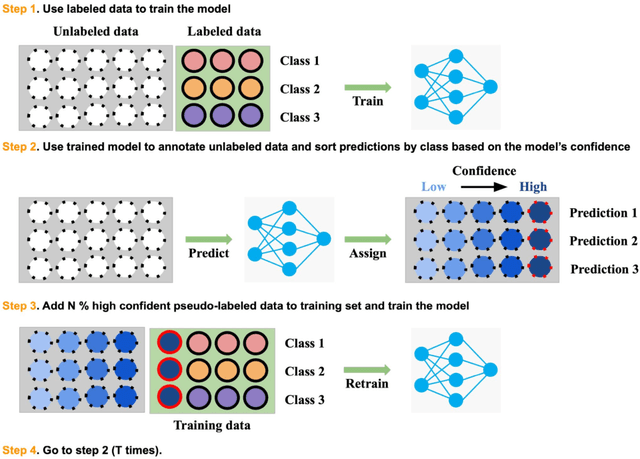
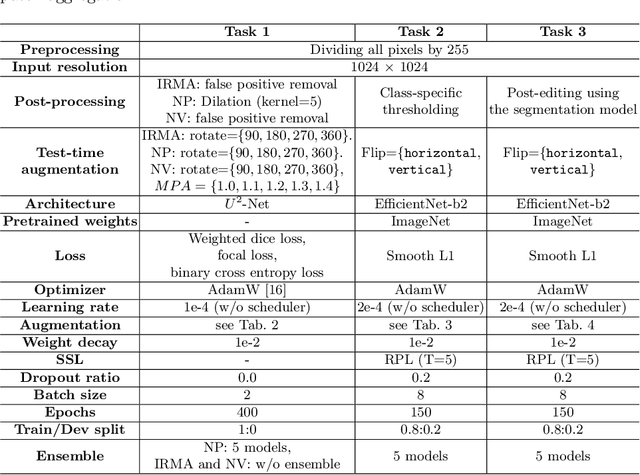
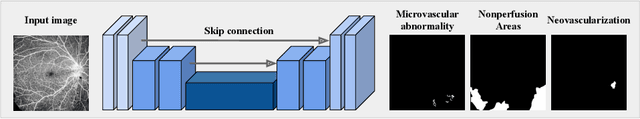
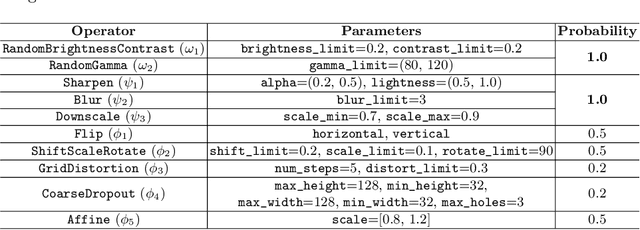
Abstract:Recently, diabetic retinopathy (DR) screening utilizing ultra-wide optical coherence tomography angiography (UW-OCTA) has been used in clinical practices to detect signs of early DR. However, developing a deep learning-based DR analysis system using UW-OCTA images is not trivial due to the difficulty of data collection and the absence of public datasets. By realistic constraints, a model trained on small datasets may obtain sub-par performance. Therefore, to help ophthalmologists be less confused about models' incorrect decisions, the models should be robust even in data scarcity settings. To address the above practical challenging, we present a comprehensive empirical study for DR analysis tasks, including lesion segmentation, image quality assessment, and DR grading. For each task, we introduce a robust training scheme by leveraging ensemble learning, data augmentation, and semi-supervised learning. Furthermore, we propose reliable pseudo labeling that excludes uncertain pseudo-labels based on the model's confidence scores to reduce the negative effect of noisy pseudo-labels. By exploiting the proposed approaches, we achieved 1st place in the Diabetic Retinopathy Analysis Challenge.
ADAM Challenge: Detecting Age-related Macular Degeneration from Fundus Images
Feb 18, 2022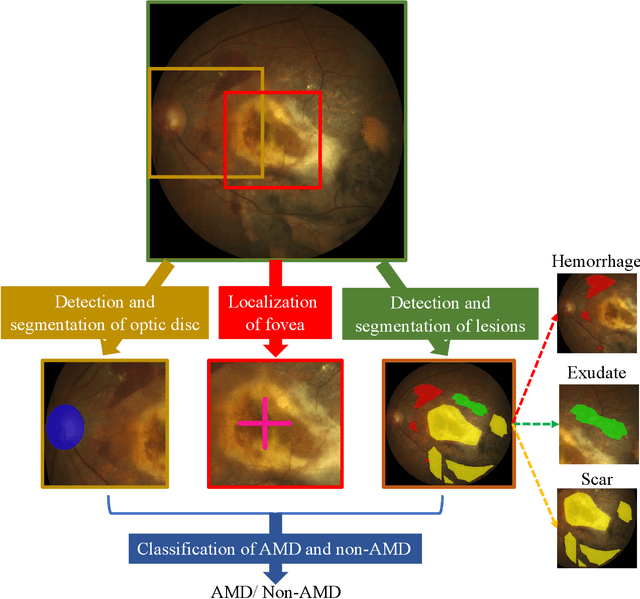
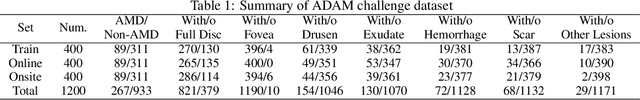
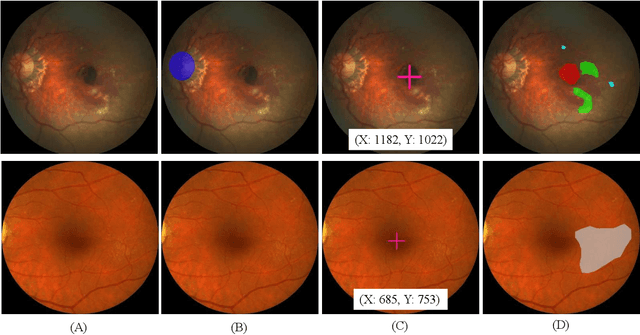
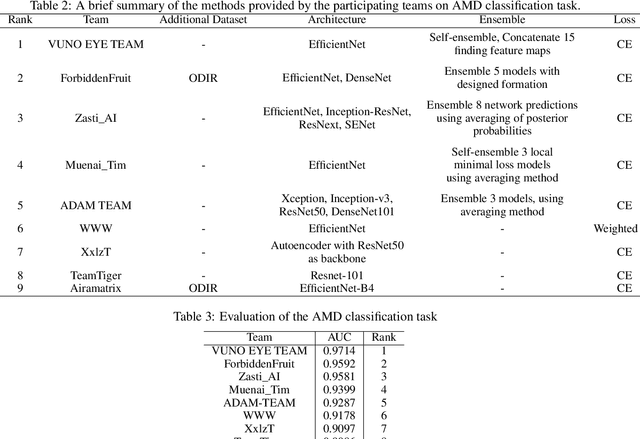
Abstract:Age-related macular degeneration (AMD) is the leading cause of visual impairment among elderly in the world. Early detection of AMD is of great importance as the vision loss caused by AMD is irreversible and permanent. Color fundus photography is the most cost-effective imaging modality to screen for retinal disorders. \textcolor{red}{Recently, some algorithms based on deep learning had been developed for fundus image analysis and automatic AMD detection. However, a comprehensive annotated dataset and a standard evaluation benchmark are still missing.} To deal with this issue, we set up the Automatic Detection challenge on Age-related Macular degeneration (ADAM) for the first time, held as a satellite event of the ISBI 2020 conference. The ADAM challenge consisted of four tasks which cover the main topics in detecting AMD from fundus images, including classification of AMD, detection and segmentation of optic disc, localization of fovea, and detection and segmentation of lesions. The ADAM challenge has released a comprehensive dataset of 1200 fundus images with the category labels of AMD, the pixel-wise segmentation masks of the full optic disc and lesions (drusen, exudate, hemorrhage, scar, and other), as well as the location coordinates of the macular fovea. A uniform evaluation framework has been built to make a fair comparison of different models. During the ADAM challenge, 610 results were submitted for online evaluation, and finally, 11 teams participated in the onsite challenge. This paper introduces the challenge, dataset, and evaluation methods, as well as summarizes the methods and analyzes the results of the participating teams of each task. In particular, we observed that ensembling strategy and clinical prior knowledge can better improve the performances of the deep learning models.
Synthesis of Brain Tumor MR Images for Learning Data Augmentation
Mar 17, 2020



Abstract:Medical image analysis using deep neural networks has been actively studied. Deep neural networks are trained by learning data. For accurate training of deep neural networks, the learning data should be sufficient, of good quality, and should have a generalized property. However, in medical images, it is difficult to acquire sufficient patient data because of the difficulty of patient recruitment, the burden of annotation of lesions by experts, and the invasion of patients' privacy. In comparison, the medical images of healthy volunteers can be easily acquired. Using healthy brain images, the proposed method synthesizes multi-contrast magnetic resonance images of brain tumors. Because tumors have complex features, the proposed method simplifies them into concentric circles that are easily controllable. Then it converts the concentric circles into various realistic shapes of tumors through deep neural networks. Because numerous healthy brain images are easily available, our method can synthesize a huge number of the brain tumor images with various concentric circles. We performed qualitative and quantitative analysis to assess the usefulness of augmented data from the proposed method. Intuitive and interesting experimental results are available online at https://github.com/KSH0660/BrainTumor
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge