Stavros Gerolymatos
On Background Bias of Post-Hoc Concept Embeddings in Computer Vision DNNs
Apr 11, 2025Abstract:The thriving research field of concept-based explainable artificial intelligence (C-XAI) investigates how human-interpretable semantic concepts embed in the latent spaces of deep neural networks (DNNs). Post-hoc approaches therein use a set of examples to specify a concept, and determine its embeddings in DNN latent space using data driven techniques. This proved useful to uncover biases between different target (foreground or concept) classes. However, given that the background is mostly uncontrolled during training, an important question has been left unattended so far: Are/to what extent are state-of-the-art, data-driven post-hoc C-XAI approaches themselves prone to biases with respect to their backgrounds? E.g., wild animals mostly occur against vegetation backgrounds, and they seldom appear on roads. Even simple and robust C-XAI methods might abuse this shortcut for enhanced performance. A dangerous performance degradation of the concept-corner cases of animals on the road could thus remain undiscovered. This work validates and thoroughly confirms that established Net2Vec-based concept segmentation techniques frequently capture background biases, including alarming ones, such as underperformance on road scenes. For the analysis, we compare 3 established techniques from the domain of background randomization on >50 concepts from 2 datasets, and 7 diverse DNN architectures. Our results indicate that even low-cost setups can provide both valuable insight and improved background robustness.
Cluster Exploration using Informative Manifold Projections
Sep 26, 2023



Abstract:Dimensionality reduction (DR) is one of the key tools for the visual exploration of high-dimensional data and uncovering its cluster structure in two- or three-dimensional spaces. The vast majority of DR methods in the literature do not take into account any prior knowledge a practitioner may have regarding the dataset under consideration. We propose a novel method to generate informative embeddings which not only factor out the structure associated with different kinds of prior knowledge but also aim to reveal any remaining underlying structure. To achieve this, we employ a linear combination of two objectives: firstly, contrastive PCA that discounts the structure associated with the prior information, and secondly, kurtosis projection pursuit which ensures meaningful data separation in the obtained embeddings. We formulate this task as a manifold optimization problem and validate it empirically across a variety of datasets considering three distinct types of prior knowledge. Lastly, we provide an automated framework to perform iterative visual exploration of high-dimensional data.
SHREC 2021: Classification in cryo-electron tomograms
Mar 18, 2022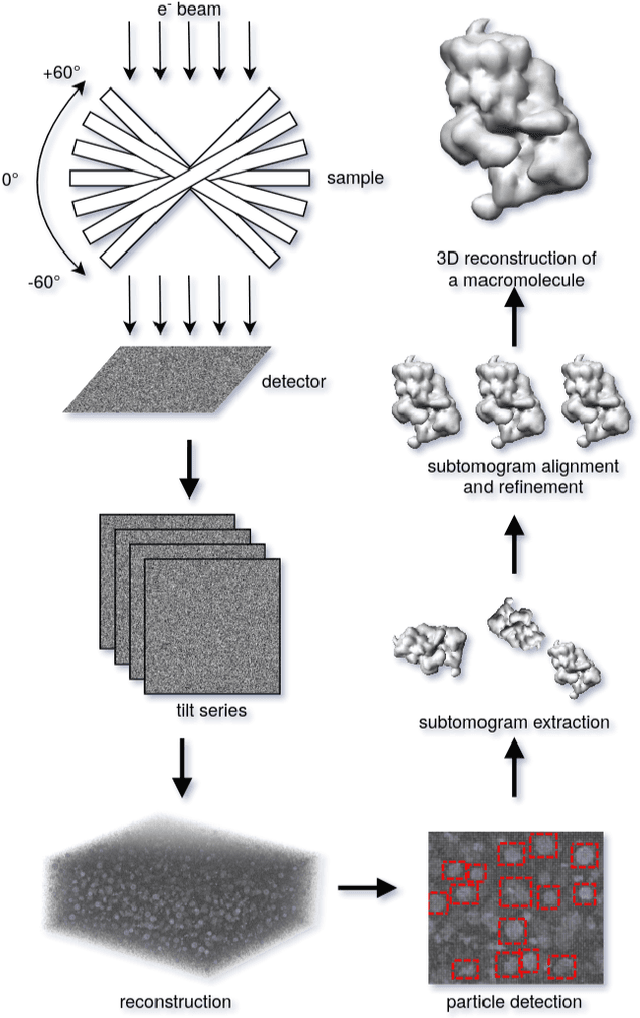
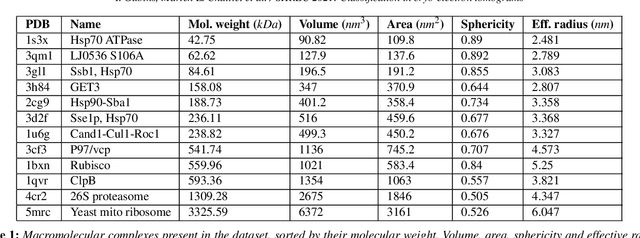
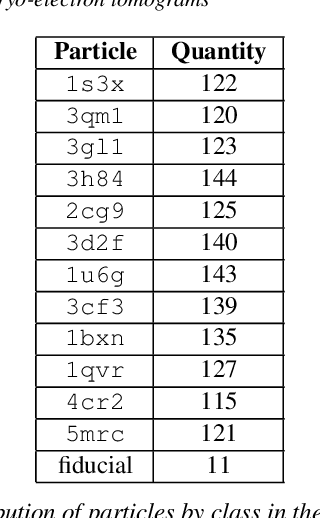
Abstract:Cryo-electron tomography (cryo-ET) is an imaging technique that allows three-dimensional visualization of macro-molecular assemblies under near-native conditions. Cryo-ET comes with a number of challenges, mainly low signal-to-noise and inability to obtain images from all angles. Computational methods are key to analyze cryo-electron tomograms. To promote innovation in computational methods, we generate a novel simulated dataset to benchmark different methods of localization and classification of biological macromolecules in tomograms. Our publicly available dataset contains ten tomographic reconstructions of simulated cell-like volumes. Each volume contains twelve different types of complexes, varying in size, function and structure. In this paper, we have evaluated seven different methods of finding and classifying proteins. Seven research groups present results obtained with learning-based methods and trained on the simulated dataset, as well as a baseline template matching (TM), a traditional method widely used in cryo-ET research. We show that learning-based approaches can achieve notably better localization and classification performance than TM. We also experimentally confirm that there is a negative relationship between particle size and performance for all methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge