Emmanuel Moebel
Normalization-Equivariant Neural Networks with Application to Image Denoising
Jun 08, 2023Abstract:In many information processing systems, it may be desirable to ensure that any change of the input, whether by shifting or scaling, results in a corresponding change in the system response. While deep neural networks are gradually replacing all traditional automatic processing methods, they surprisingly do not guarantee such normalization-equivariance (scale + shift) property, which can be detrimental in many applications. To address this issue, we propose a methodology for adapting existing neural networks so that normalization-equivariance holds by design. Our main claim is that not only ordinary convolutional layers, but also all activation functions, including the ReLU (rectified linear unit), which are applied element-wise to the pre-activated neurons, should be completely removed from neural networks and replaced by better conditioned alternatives. To this end, we introduce affine-constrained convolutions and channel-wise sort pooling layers as surrogates and show that these two architectural modifications do preserve normalization-equivariance without loss of performance. Experimental results in image denoising show that normalization-equivariant neural networks, in addition to their better conditioning, also provide much better generalization across noise levels.
SHREC 2021: Classification in cryo-electron tomograms
Mar 18, 2022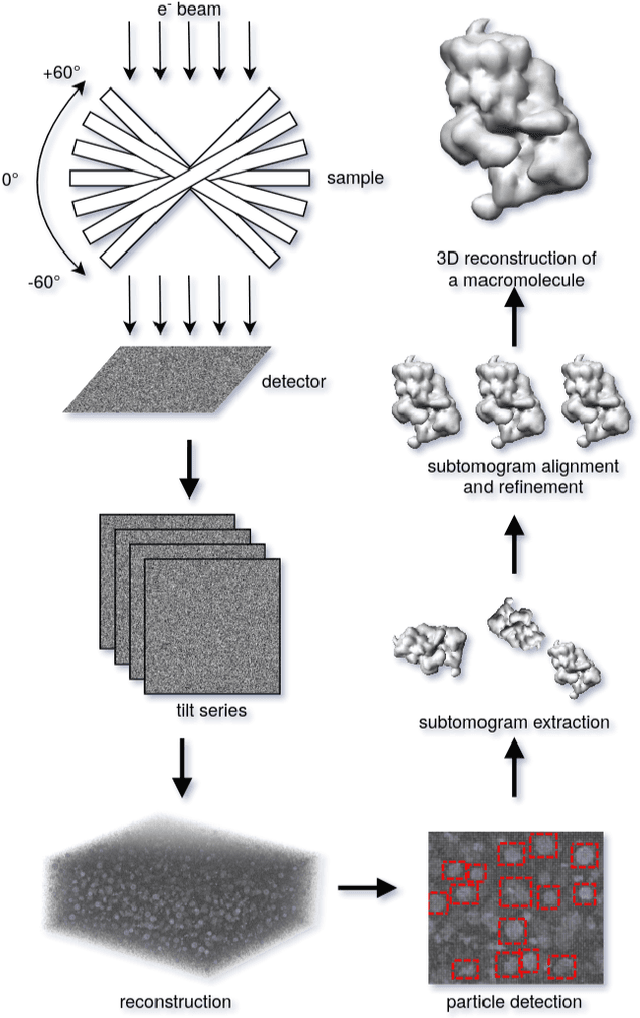
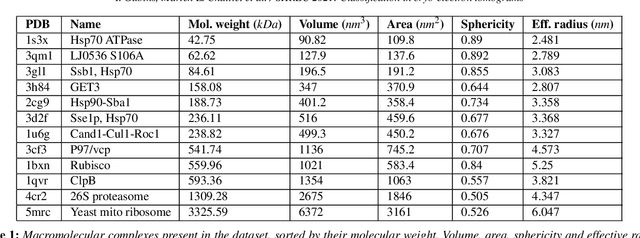
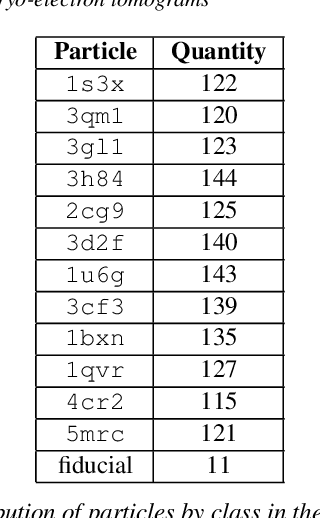
Abstract:Cryo-electron tomography (cryo-ET) is an imaging technique that allows three-dimensional visualization of macro-molecular assemblies under near-native conditions. Cryo-ET comes with a number of challenges, mainly low signal-to-noise and inability to obtain images from all angles. Computational methods are key to analyze cryo-electron tomograms. To promote innovation in computational methods, we generate a novel simulated dataset to benchmark different methods of localization and classification of biological macromolecules in tomograms. Our publicly available dataset contains ten tomographic reconstructions of simulated cell-like volumes. Each volume contains twelve different types of complexes, varying in size, function and structure. In this paper, we have evaluated seven different methods of finding and classifying proteins. Seven research groups present results obtained with learning-based methods and trained on the simulated dataset, as well as a baseline template matching (TM), a traditional method widely used in cryo-ET research. We show that learning-based approaches can achieve notably better localization and classification performance than TM. We also experimentally confirm that there is a negative relationship between particle size and performance for all methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge