Shiyun Xie
SuperGS: Consistent and Detailed 3D Super-Resolution Scene Reconstruction via Gaussian Splatting
May 24, 2025Abstract:Recently, 3D Gaussian Splatting (3DGS) has excelled in novel view synthesis (NVS) with its real-time rendering capabilities and superior quality. However, it encounters challenges for high-resolution novel view synthesis (HRNVS) due to the coarse nature of primitives derived from low-resolution input views. To address this issue, we propose SuperGS, an expansion of Scaffold-GS designed with a two-stage coarse-to-fine training framework. In the low-resolution stage, we introduce a latent feature field to represent the low-resolution scene, which serves as both the initialization and foundational information for super-resolution optimization. In the high-resolution stage, we propose a multi-view consistent densification strategy that backprojects high-resolution depth maps based on error maps and employs a multi-view voting mechanism, mitigating ambiguities caused by multi-view inconsistencies in the pseudo labels provided by 2D prior models while avoiding Gaussian redundancy. Furthermore, we model uncertainty through variational feature learning and use it to guide further scene representation refinement and adjust the supervisory effect of pseudo-labels, ensuring consistent and detailed scene reconstruction. Extensive experiments demonstrate that SuperGS outperforms state-of-the-art HRNVS methods on both forward-facing and 360-degree datasets.
SuperGS: Super-Resolution 3D Gaussian Splatting via Latent Feature Field and Gradient-guided Splitting
Oct 03, 2024Abstract:Recently, 3D Gaussian Splatting (3DGS) has exceled in novel view synthesis with its real-time rendering capabilities and superior quality. However, it faces challenges for high-resolution novel view synthesis (HRNVS) due to the coarse nature of primitives derived from low-resolution input views. To address this issue, we propose Super-Resolution 3DGS (SuperGS), which is an expansion of 3DGS designed with a two-stage coarse-to-fine training framework, utilizing pretrained low-resolution scene representation as an initialization for super-resolution optimization. Moreover, we introduce Multi-resolution Feature Gaussian Splatting (MFGS) to incorporates a latent feature field for flexible feature sampling and Gradient-guided Selective Splitting (GSS) for effective Gaussian upsampling. By integrating these strategies within the coarse-to-fine framework ensure both high fidelity and memory efficiency. Extensive experiments demonstrate that SuperGS surpasses state-of-the-art HRNVS methods on challenging real-world datasets using only low-resolution inputs.
EMERGE: Integrating RAG for Improved Multimodal EHR Predictive Modeling
May 27, 2024



Abstract:The integration of multimodal Electronic Health Records (EHR) data has notably advanced clinical predictive capabilities. However, current models that utilize clinical notes and multivariate time-series EHR data often lack the necessary medical context for precise clinical tasks. Previous methods using knowledge graphs (KGs) primarily focus on structured knowledge extraction. To address this, we propose EMERGE, a Retrieval-Augmented Generation (RAG) driven framework aimed at enhancing multimodal EHR predictive modeling. Our approach extracts entities from both time-series data and clinical notes by prompting Large Language Models (LLMs) and aligns them with professional PrimeKG to ensure consistency. Beyond triplet relationships, we include entities' definitions and descriptions to provide richer semantics. The extracted knowledge is then used to generate task-relevant summaries of patients' health statuses. These summaries are fused with other modalities utilizing an adaptive multimodal fusion network with cross-attention. Extensive experiments on the MIMIC-III and MIMIC-IV datasets for in-hospital mortality and 30-day readmission tasks demonstrate the superior performance of the EMERGE framework compared to baseline models. Comprehensive ablation studies and analyses underscore the efficacy of each designed module and the framework's robustness to data sparsity. EMERGE significantly enhances the use of multimodal EHR data in healthcare, bridging the gap with nuanced medical contexts crucial for informed clinical predictions.
REALM: RAG-Driven Enhancement of Multimodal Electronic Health Records Analysis via Large Language Models
Feb 10, 2024Abstract:The integration of multimodal Electronic Health Records (EHR) data has significantly improved clinical predictive capabilities. Leveraging clinical notes and multivariate time-series EHR, existing models often lack the medical context relevent to clinical tasks, prompting the incorporation of external knowledge, particularly from the knowledge graph (KG). Previous approaches with KG knowledge have primarily focused on structured knowledge extraction, neglecting unstructured data modalities and semantic high dimensional medical knowledge. In response, we propose REALM, a Retrieval-Augmented Generation (RAG) driven framework to enhance multimodal EHR representations that address these limitations. Firstly, we apply Large Language Model (LLM) to encode long context clinical notes and GRU model to encode time-series EHR data. Secondly, we prompt LLM to extract task-relevant medical entities and match entities in professionally labeled external knowledge graph (PrimeKG) with corresponding medical knowledge. By matching and aligning with clinical standards, our framework eliminates hallucinations and ensures consistency. Lastly, we propose an adaptive multimodal fusion network to integrate extracted knowledge with multimodal EHR data. Our extensive experiments on MIMIC-III mortality and readmission tasks showcase the superior performance of our REALM framework over baselines, emphasizing the effectiveness of each module. REALM framework contributes to refining the use of multimodal EHR data in healthcare and bridging the gap with nuanced medical context essential for informed clinical predictions.
Leveraging Prototype Patient Representations with Feature-Missing-Aware Calibration to Mitigate EHR Data Sparsity
Sep 17, 2023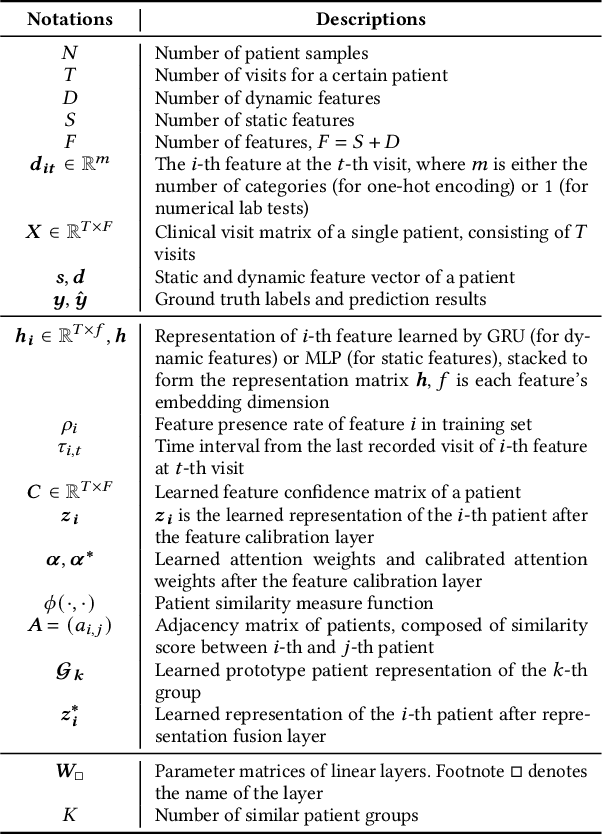
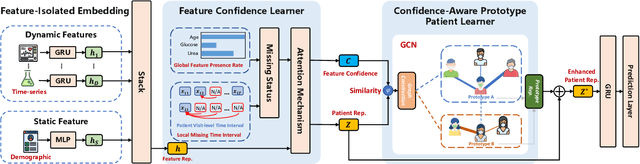
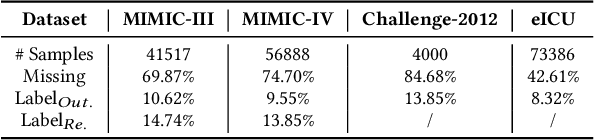
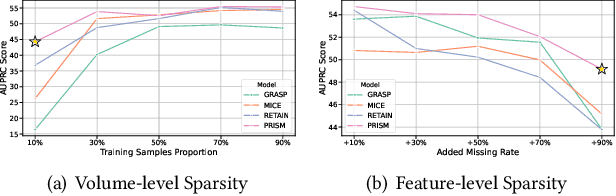
Abstract:Electronic Health Record (EHR) data frequently exhibits sparse characteristics, posing challenges for predictive modeling. Current direct imputation such as matrix imputation approaches hinge on referencing analogous rows or columns to complete raw missing data and do not differentiate between imputed and actual values. As a result, models may inadvertently incorporate irrelevant or deceptive information with respect to the prediction objective, thereby compromising the efficacy of downstream performance. While some methods strive to recalibrate or augment EHR embeddings after direct imputation, they often mistakenly prioritize imputed features. This misprioritization can introduce biases or inaccuracies into the model. To tackle these issues, our work resorts to indirect imputation, where we leverage prototype representations from similar patients to obtain a denser embedding. Recognizing the limitation that missing features are typically treated the same as present ones when measuring similar patients, our approach designs a feature confidence learner module. This module is sensitive to the missing feature status, enabling the model to better judge the reliability of each feature. Moreover, we propose a novel patient similarity metric that takes feature confidence into account, ensuring that evaluations are not based merely on potentially inaccurate imputed values. Consequently, our work captures dense prototype patient representations with feature-missing-aware calibration process. Comprehensive experiments demonstrate that designed model surpasses established EHR-focused models with a statistically significant improvement on MIMIC-III and MIMIC-IV datasets in-hospital mortality outcome prediction task. The code is publicly available at \url{https://github.com/yhzhu99/SparseEHR} to assure the reproducibility.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge