Shir Ashury-Tahan
Robustness as an Emergent Property of Task Performance
Feb 03, 2026Abstract:Robustness is often regarded as a critical future challenge for real-world applications, where stability is essential. However, as models often learn tasks in a similar order, we hypothesize that easier tasks will be easier regardless of how they are presented to the model. Indeed, in this paper, we show that as models approach high performance on a task, robustness is effectively achieved. Through an empirical analysis of multiple models across diverse datasets and configurations (e.g., paraphrases, different temperatures), we find a strong positive correlation. Moreover, we find that robustness is primarily driven by task-specific competence rather than inherent model-level properties, challenging current approaches that treat robustness as an independent capability. Thus, from a high-level perspective, we may expect that as new tasks saturate, model robustness on these tasks will emerge accordingly. For researchers, this implies that explicit efforts to measure and improve robustness may warrant reduced emphasis, as such robustness is likely to develop alongside performance gains. For practitioners, it acts as a sign that indeed the tasks that the literature deals with are unreliable, but on easier past tasks, the models are reliable and ready for real-world deployment.
ErrorMap and ErrorAtlas: Charting the Failure Landscape of Large Language Models
Jan 22, 2026Abstract:Large Language Models (LLM) benchmarks tell us when models fail, but not why they fail. A wrong answer on a reasoning dataset may stem from formatting issues, calculation errors, or dataset noise rather than weak reasoning. Without disentangling such causes, benchmarks remain incomplete and cannot reliably guide model improvement. We introduce ErrorMap, the first method to chart the sources of LLM failure. It extracts a model's unique "failure signature", clarifies what benchmarks measure, and broadens error identification to reduce blind spots. This helps developers debug models, aligns benchmark goals with outcomes, and supports informed model selection. ErrorMap works on any model or dataset with the same logic. Applying our method to 35 datasets and 83 models we generate ErrorAtlas, a taxonomy of model errors, revealing recurring failure patterns. ErrorAtlas highlights error types that are currently underexplored in LLM research, such as omissions of required details in the output and question misinterpretation. By shifting focus from where models succeed to why they fail, ErrorMap and ErrorAtlas enable advanced evaluation - one that exposes hidden weaknesses and directs progress. Unlike success, typically measured by task-level metrics, our approach introduces a deeper evaluation layer that can be applied globally across models and tasks, offering richer insights into model behavior and limitations. We make the taxonomy and code publicly available with plans to periodically update ErrorAtlas as new benchmarks and models emerge.
The Mighty ToRR: A Benchmark for Table Reasoning and Robustness
Feb 26, 2025



Abstract:Despite its real-world significance, model performance on tabular data remains underexplored, leaving uncertainty about which model to rely on and which prompt configuration to adopt. To address this gap, we create ToRR, a benchmark for Table Reasoning and Robustness, that measures model performance and robustness on table-related tasks. The benchmark includes 10 datasets that cover different types of table reasoning capabilities across varied domains. ToRR goes beyond model performance rankings, and is designed to reflect whether models can handle tabular data consistently and robustly, across a variety of common table representation formats. We present a leaderboard as well as comprehensive analyses of the results of leading models over ToRR. Our results reveal a striking pattern of brittle model behavior, where even strong models are unable to perform robustly on tabular data tasks. Although no specific table format leads to consistently better performance, we show that testing over multiple formats is crucial for reliably estimating model capabilities. Moreover, we show that the reliability boost from testing multiple prompts can be equivalent to adding more test examples. Overall, our findings show that table understanding and reasoning tasks remain a significant challenge.
Data-driven Coreference-based Ontology Building
Oct 22, 2024Abstract:While coreference resolution is traditionally used as a component in individual document understanding, in this work we take a more global view and explore what can we learn about a domain from the set of all document-level coreference relations that are present in a large corpus. We derive coreference chains from a corpus of 30 million biomedical abstracts and construct a graph based on the string phrases within these chains, establishing connections between phrases if they co-occur within the same coreference chain. We then use the graph structure and the betweeness centrality measure to distinguish between edges denoting hierarchy, identity and noise, assign directionality to edges denoting hierarchy, and split nodes (strings) that correspond to multiple distinct concepts. The result is a rich, data-driven ontology over concepts in the biomedical domain, parts of which overlaps significantly with human-authored ontologies. We release the coreference chains and resulting ontology under a creative-commons license, along with the code.
Label-Efficient Model Selection for Text Generation
Feb 12, 2024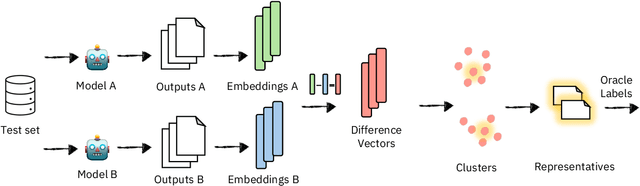

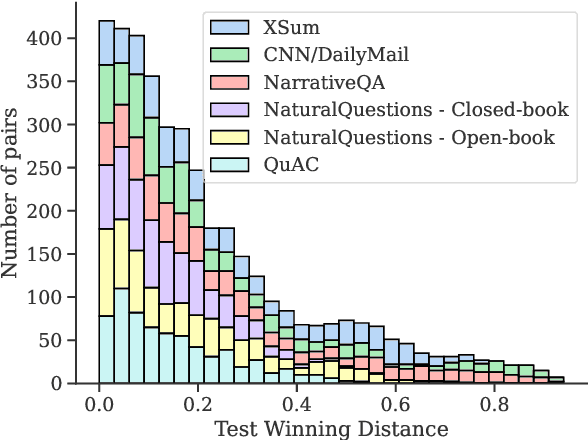
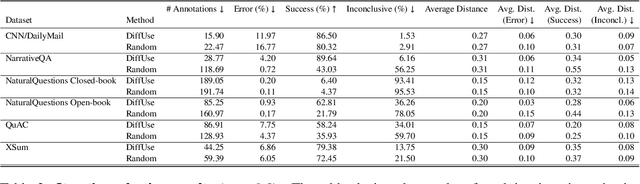
Abstract:Model selection for a given target task can be costly, as it may entail extensive annotation of the quality of outputs of different models. We introduce DiffUse, an efficient method to make an informed decision between candidate text generation models. DiffUse reduces the required amount of preference annotations, thus saving valuable time and resources in performing evaluation. DiffUse intelligently selects instances by clustering embeddings that represent the semantic differences between model outputs. Thus, it is able to identify a subset of examples that are more informative for preference decisions. Our method is model-agnostic, and can be applied to any text generation model. Moreover, we propose a practical iterative approach for dynamically determining how many instances to annotate. In a series of experiments over hundreds of model pairs, we demonstrate that DiffUse can dramatically reduce the required number of annotations -- by up to 75% -- while maintaining high evaluation reliability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge