Runyuan Bao
Deep Learning in Image Classification: Evaluating VGG19's Performance on Complex Visual Data
Dec 29, 2024



Abstract:This study aims to explore the automatic classification method of pneumonia X-ray images based on VGG19 deep convolutional neural network, and evaluate its application effect in pneumonia diagnosis by comparing with classic models such as SVM, XGBoost, MLP, and ResNet50. The experimental results show that VGG19 performs well in multiple indicators such as accuracy (92%), AUC (0.95), F1 score (0.90) and recall rate (0.87), which is better than other comparison models, especially in image feature extraction and classification accuracy. Although ResNet50 performs well in some indicators, it is slightly inferior to VGG19 in recall rate and F1 score. Traditional machine learning models SVM and XGBoost are obviously limited in image classification tasks, especially in complex medical image analysis tasks, and their performance is relatively mediocre. The research results show that deep learning, especially convolutional neural networks, have significant advantages in medical image classification tasks, especially in pneumonia X-ray image analysis, and can provide efficient and accurate automatic diagnosis support. This research provides strong technical support for the early detection of pneumonia and the development of automated diagnosis systems and also lays the foundation for further promoting the application and development of automated medical image processing technology.
Accurate Medical Named Entity Recognition Through Specialized NLP Models
Dec 11, 2024


Abstract:This study evaluated the effect of BioBERT in medical text processing for the task of medical named entity recognition. Through comparative experiments with models such as BERT, ClinicalBERT, SciBERT, and BlueBERT, the results showed that BioBERT achieved the best performance in both precision and F1 score, verifying its applicability and superiority in the medical field. BioBERT enhances its ability to understand professional terms and complex medical texts through pre-training on biomedical data, providing a powerful tool for medical information extraction and clinical decision support. The study also explored the privacy and compliance challenges of BioBERT when processing medical data, and proposed future research directions for combining other medical-specific models to improve generalization and robustness. With the development of deep learning technology, the potential of BioBERT in application fields such as intelligent medicine, personalized treatment, and disease prediction will be further expanded. Future research can focus on the real-time and interpretability of the model to promote its widespread application in the medical field.
Few-Shot Learning with Adaptive Weight Masking in Conditional GANs
Dec 04, 2024



Abstract:Deep learning has revolutionized various fields, yet its efficacy is hindered by overfitting and the requirement of extensive annotated data, particularly in few-shot learning scenarios where limited samples are available. This paper introduces a novel approach to few-shot learning by employing a Residual Weight Masking Conditional Generative Adversarial Network (RWM-CGAN) for data augmentation. The proposed model integrates residual units within the generator to enhance network depth and sample quality, coupled with a weight mask regularization technique in the discriminator to improve feature learning from small-sample categories. This method addresses the core issues of robustness and generalization in few-shot learning by providing a controlled and clear augmentation of the sample space. Extensive experiments demonstrate that RWM-CGAN not only expands the sample space effectively but also enriches the diversity and quality of generated samples, leading to significant improvements in detection and classification accuracy on public datasets. The paper contributes to the advancement of few-shot learning by offering a practical solution to the challenges posed by data scarcity and the need for rapid generalization to new tasks or categories.
Deep Learning for Medical Text Processing: BERT Model Fine-Tuning and Comparative Study
Oct 28, 2024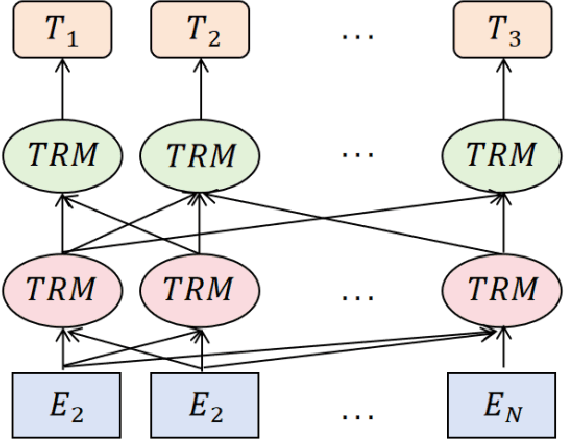
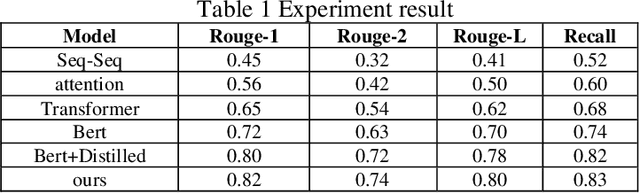
Abstract:This paper proposes a medical literature summary generation method based on the BERT model to address the challenges brought by the current explosion of medical information. By fine-tuning and optimizing the BERT model, we develop an efficient summary generation system that can quickly extract key information from medical literature and generate coherent, accurate summaries. In the experiment, we compared various models, including Seq-Seq, Attention, Transformer, and BERT, and demonstrated that the improved BERT model offers significant advantages in the Rouge and Recall metrics. Furthermore, the results of this study highlight the potential of knowledge distillation techniques to further enhance model performance. The system has demonstrated strong versatility and efficiency in practical applications, offering a reliable tool for the rapid screening and analysis of medical literature.
Optimizing Retrieval-Augmented Generation with Elasticsearch for Enhanced Question-Answering Systems
Oct 18, 2024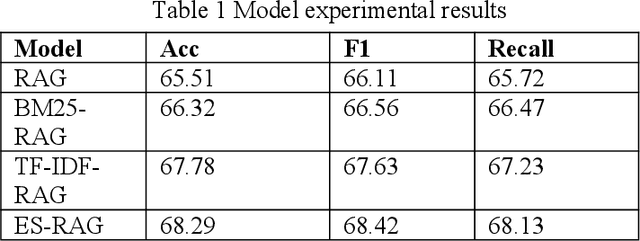
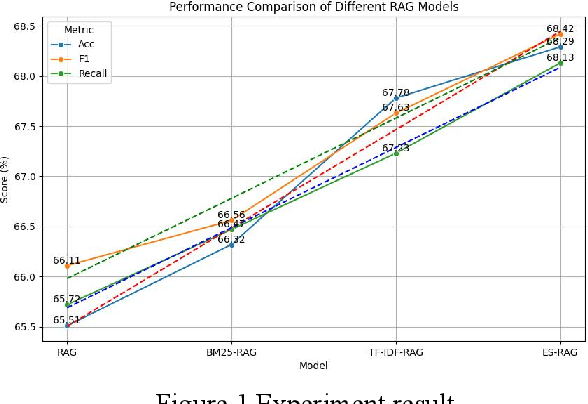
Abstract:This study aims to improve the accuracy and quality of large-scale language models (LLMs) in answering questions by integrating Elasticsearch into the Retrieval Augmented Generation (RAG) framework. The experiment uses the Stanford Question Answering Dataset (SQuAD) version 2.0 as the test dataset and compares the performance of different retrieval methods, including traditional methods based on keyword matching or semantic similarity calculation, BM25-RAG and TF-IDF- RAG, and the newly proposed ES-RAG scheme. The results show that ES-RAG not only has obvious advantages in retrieval efficiency but also performs well in key indicators such as accuracy, which is 0.51 percentage points higher than TF-IDF-RAG. In addition, Elasticsearch's powerful search capabilities and rich configuration options enable the entire question-answering system to better handle complex queries and provide more flexible and efficient responses based on the diverse needs of users. Future research directions can further explore how to optimize the interaction mechanism between Elasticsearch and LLM, such as introducing higher-level semantic understanding and context-awareness capabilities, to achieve a more intelligent and humanized question-answering experience.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge