Hanchao Zhang
Adaptive Transformer Attention and Multi-Scale Fusion for Spine 3D Segmentation
Mar 17, 2025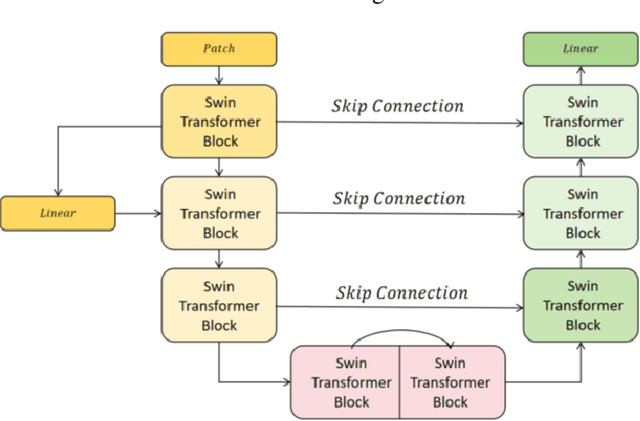
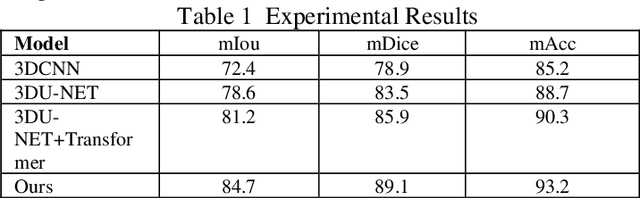
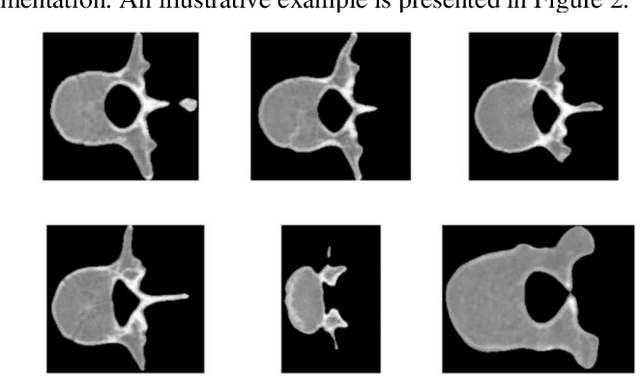
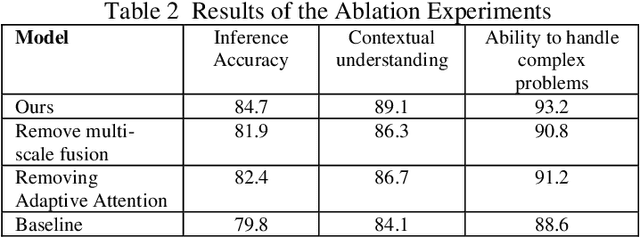
Abstract:This study proposes a 3D semantic segmentation method for the spine based on the improved SwinUNETR to improve segmentation accuracy and robustness. Aiming at the complex anatomical structure of spinal images, this paper introduces a multi-scale fusion mechanism to enhance the feature extraction capability by using information of different scales, thereby improving the recognition accuracy of the model for the target area. In addition, the introduction of the adaptive attention mechanism enables the model to dynamically adjust the attention to the key area, thereby optimizing the boundary segmentation effect. The experimental results show that compared with 3D CNN, 3D U-Net, and 3D U-Net + Transformer, the model of this study has achieved significant improvements in mIoU, mDice, and mAcc indicators, and has better segmentation performance. The ablation experiment further verifies the effectiveness of the proposed improved method, proving that multi-scale fusion and adaptive attention mechanism have a positive effect on the segmentation task. Through the visualization analysis of the inference results, the model can better restore the real anatomical structure of the spinal image. Future research can further optimize the Transformer structure and expand the data scale to improve the generalization ability of the model. This study provides an efficient solution for the task of medical image segmentation, which is of great significance to intelligent medical image analysis.
Multi-Scale Transformer Architecture for Accurate Medical Image Classification
Feb 10, 2025



Abstract:This study introduces an AI-driven skin lesion classification algorithm built on an enhanced Transformer architecture, addressing the challenges of accuracy and robustness in medical image analysis. By integrating a multi-scale feature fusion mechanism and refining the self-attention process, the model effectively extracts both global and local features, enhancing its ability to detect lesions with ambiguous boundaries and intricate structures. Performance evaluation on the ISIC 2017 dataset demonstrates that the improved Transformer surpasses established AI models, including ResNet50, VGG19, ResNext, and Vision Transformer, across key metrics such as accuracy, AUC, F1-Score, and Precision. Grad-CAM visualizations further highlight the interpretability of the model, showcasing strong alignment between the algorithm's focus areas and actual lesion sites. This research underscores the transformative potential of advanced AI models in medical imaging, paving the way for more accurate and reliable diagnostic tools. Future work will explore the scalability of this approach to broader medical imaging tasks and investigate the integration of multimodal data to enhance AI-driven diagnostic frameworks for intelligent healthcare.
Accurate Medical Named Entity Recognition Through Specialized NLP Models
Dec 11, 2024


Abstract:This study evaluated the effect of BioBERT in medical text processing for the task of medical named entity recognition. Through comparative experiments with models such as BERT, ClinicalBERT, SciBERT, and BlueBERT, the results showed that BioBERT achieved the best performance in both precision and F1 score, verifying its applicability and superiority in the medical field. BioBERT enhances its ability to understand professional terms and complex medical texts through pre-training on biomedical data, providing a powerful tool for medical information extraction and clinical decision support. The study also explored the privacy and compliance challenges of BioBERT when processing medical data, and proposed future research directions for combining other medical-specific models to improve generalization and robustness. With the development of deep learning technology, the potential of BioBERT in application fields such as intelligent medicine, personalized treatment, and disease prediction will be further expanded. Future research can focus on the real-time and interpretability of the model to promote its widespread application in the medical field.
Pattern Integration and Enhancement Vision Transformer for Self-Supervised Learning in Remote Sensing
Nov 09, 2024



Abstract:Recent self-supervised learning (SSL) methods have demonstrated impressive results in learning visual representations from unlabeled remote sensing images. However, most remote sensing images predominantly consist of scenographic scenes containing multiple ground objects without explicit foreground targets, which limits the performance of existing SSL methods that focus on foreground targets. This raises the question: Is there a method that can automatically aggregate similar objects within scenographic remote sensing images, thereby enabling models to differentiate knowledge embedded in various geospatial patterns for improved feature representation? In this work, we present the Pattern Integration and Enhancement Vision Transformer (PIEViT), a novel self-supervised learning framework designed specifically for remote sensing imagery. PIEViT utilizes a teacher-student architecture to address both image-level and patch-level tasks. It employs the Geospatial Pattern Cohesion (GPC) module to explore the natural clustering of patches, enhancing the differentiation of individual features. The Feature Integration Projection (FIP) module further refines masked token reconstruction using geospatially clustered patches. We validated PIEViT across multiple downstream tasks, including object detection, semantic segmentation, and change detection. Experiments demonstrated that PIEViT enhances the representation of internal patch features, providing significant improvements over existing self-supervised baselines. It achieves excellent results in object detection, land cover classification, and change detection, underscoring its robustness, generalization, and transferability for remote sensing image interpretation tasks.
K-Tensors: Clustering Positive Semi-Definite Matrices
Jun 10, 2023Abstract:This paper introduces a novel self-consistency clustering algorithm (K-Tensors) designed for positive-semidefinite matrices based on their eigenstructures. As positive semi-definite matrices can be represented as ellipses or ellipsoids in $\Re^p$, $p \ge 2$, it is critical to maintain their structural information to perform effective clustering. However, traditional clustering algorithms often vectorize the matrices, resulting in a loss of essential structural information. To address this issue, we propose a distance metric that is specifically based on the structural information of positive semi-definite matrices. This distance metric enables the clustering algorithm to consider the differences between positive semi-definite matrices and their projection onto the common space spanned by a set of positive semi-definite matrices. This innovative approach to clustering positive semi-definite matrices has broad applications in several domains, including financial and biomedical research, such as analyzing functional connectivity data. By maintaining the structural information of positive semi-definite matrices, our proposed algorithm promises to cluster the positive semi-definite matrices in a more meaningful way, thereby facilitating deeper insights into the underlying data in various applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge