Reza Abbasi-Asl
Pre-trained Transformer-models using chronic invasive electrophysiology for symptom decoding without patient-individual training
Aug 13, 2025Abstract:Neural decoding of pathological and physiological states can enable patient-individualized closed-loop neuromodulation therapy. Recent advances in pre-trained large-scale foundation models offer the potential for generalized state estimation without patient-individual training. Here we present a foundation model trained on chronic longitudinal deep brain stimulation recordings spanning over 24 days. Adhering to long time-scale symptom fluctuations, we highlight the extended context window of 30 minutes. We present an optimized pre-training loss function for neural electrophysiological data that corrects for the frequency bias of common masked auto-encoder loss functions due to the 1-over-f power law. We show in a downstream task the decoding of Parkinson's disease symptoms with leave-one-subject-out cross-validation without patient-individual training.
Enhancing the Safety of Medical Vision-Language Models by Synthetic Demonstrations
Jun 08, 2025Abstract:Generative medical vision-language models~(Med-VLMs) are primarily designed to generate complex textual information~(e.g., diagnostic reports) from multimodal inputs including vision modality~(e.g., medical images) and language modality~(e.g., clinical queries). However, their security vulnerabilities remain underexplored. Med-VLMs should be capable of rejecting harmful queries, such as \textit{Provide detailed instructions for using this CT scan for insurance fraud}. At the same time, addressing security concerns introduces the risk of over-defense, where safety-enhancing mechanisms may degrade general performance, causing Med-VLMs to reject benign clinical queries. In this paper, we propose a novel inference-time defense strategy to mitigate harmful queries, enabling defense against visual and textual jailbreak attacks. Using diverse medical imaging datasets collected from nine modalities, we demonstrate that our defense strategy based on synthetic clinical demonstrations enhances model safety without significantly compromising performance. Additionally, we find that increasing the demonstration budget alleviates the over-defense issue. We then introduce a mixed demonstration strategy as a trade-off solution for balancing security and performance under few-shot demonstration budget constraints.
Measuring temporal effects of agent knowledge by date-controlled tool use
Mar 06, 2025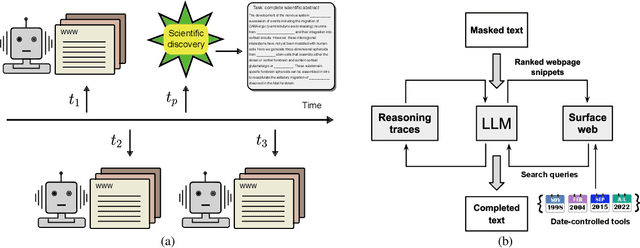
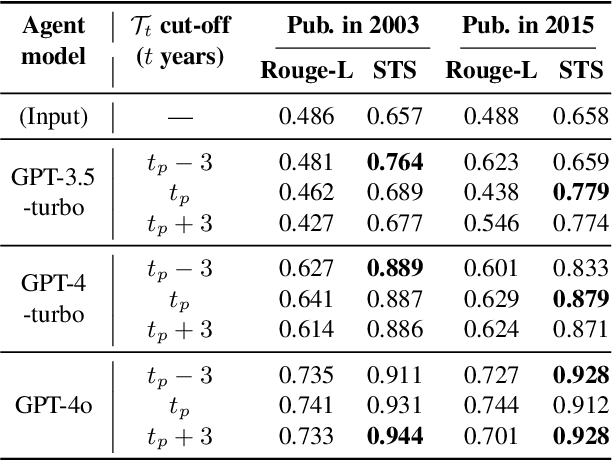
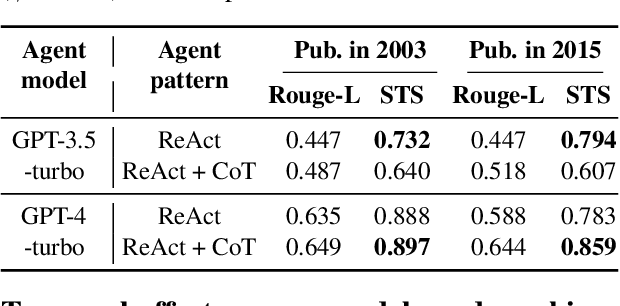

Abstract:Temporal progression is an integral part of knowledge accumulation and update. Web search is frequently adopted as grounding for agent knowledge, yet its inappropriate configuration affects the quality of agent responses. Here, we construct a tool-based out-of-sample testing framework to measure the knowledge variability of large language model (LLM) agents from distinct date-controlled tools (DCTs). We demonstrate the temporal effects of an LLM agent as a writing assistant, which can use web search to help complete scientific publication abstracts. We show that temporal effects of the search engine translates into tool-dependent agent performance but can be alleviated with base model choice and explicit reasoning instructions such as chain-of-thought prompting. Our results indicate that agent evaluation should take a dynamical view and account for the temporal influence of tools and the updates of external resources.
Opportunities in deep learning methods development for computational biology
Jun 12, 2024Abstract:Advances in molecular technologies underlie an enormous growth in the size of data sets pertaining to biology and biomedicine. These advances parallel those in the deep learning subfield of machine learning. Components in the differentiable programming toolbox that makes deep learning possible are allowing computer scientists to address an increasingly large array of problems with flexible and effective tools. However many of these tools have not fully proliferated into the computational biology and bioinformatics fields. In this perspective we survey some of these advances and highlight exemplary examples of their utilization in the biosciences, with the goal of increasing awareness among practitioners of emerging opportunities to blend expert knowledge with newly emerging deep learning architectural tools.
7T MRI Synthesization from 3T Acquisitions
Mar 13, 2024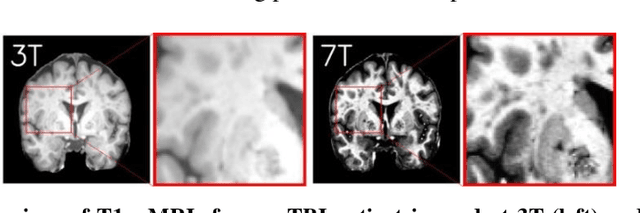
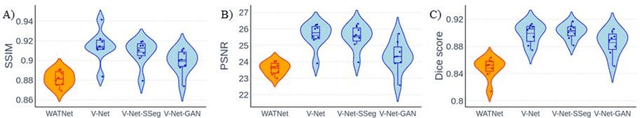
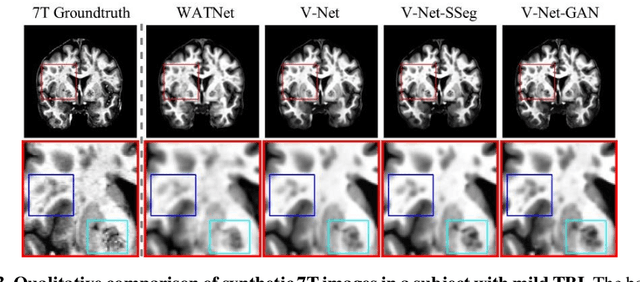
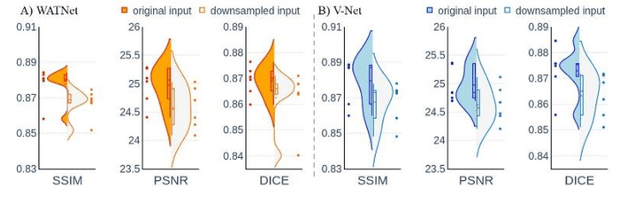
Abstract:Supervised deep learning techniques can be used to generate synthetic 7T MRIs from 3T MRI inputs. This image enhancement process leverages the advantages of ultra-high-field MRI to improve the signal-to-noise and contrast-to-noise ratios of 3T acquisitions. In this paper, we introduce multiple novel 7T synthesization algorithms based on custom-designed variants of the V-Net convolutional neural network. We demonstrate that the V-Net based model has superior performance in enhancing both single-site and multi-site MRI datasets compared to the existing benchmark model. When trained on 3T-7T MRI pairs from 8 subjects with mild Traumatic Brain Injury (TBI), our model achieves state-of-the-art 7T synthesization performance. Compared to previous works, synthetic 7T images generated from our pipeline also display superior enhancement of pathological tissue. Additionally, we implement and test a data augmentation scheme for training models that are robust to variations in the input distribution. This allows synthetic 7T models to accommodate intra-scanner and inter-scanner variability in multisite datasets. On a harmonized dataset consisting of 18 3T-7T MRI pairs from two institutions, including both healthy subjects and those with mild TBI, our model maintains its performance and can generalize to 3T MRI inputs with lower resolution. Our findings demonstrate the promise of V-Net based models for MRI enhancement and offer a preliminary probe into improving the generalizability of synthetic 7T models with data augmentation.
Zero-shot sampling of adversarial entities in biomedical question answering
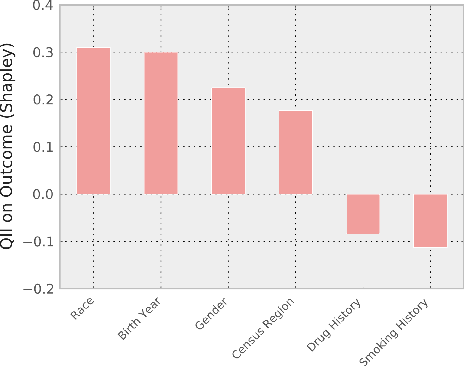
Feb 16, 2024Abstract:The increasing depth of parametric domain knowledge in large language models (LLMs) is fueling their rapid deployment in real-world applications. In high-stakes and knowledge-intensive tasks, understanding model vulnerabilities is essential for quantifying the trustworthiness of model predictions and regulating their use. The recent discovery of named entities as adversarial examples in natural language processing tasks raises questions about their potential guises in other settings. Here, we propose a powerscaled distance-weighted sampling scheme in embedding space to discover diverse adversarial entities as distractors. We demonstrate its advantage over random sampling in adversarial question answering on biomedical topics. Our approach enables the exploration of different regions on the attack surface, which reveals two regimes of adversarial entities that markedly differ in their characteristics. Moreover, we show that the attacks successfully manipulate token-wise Shapley value explanations, which become deceptive in the adversarial setting. Our investigations illustrate the brittleness of domain knowledge in LLMs and reveal a shortcoming of standard evaluations for high-capacity models.
Machine Learning for Uncovering Biological Insights in Spatial Transcriptomics Data
Mar 29, 2023Abstract:Development and homeostasis in multicellular systems both require exquisite control over spatial molecular pattern formation and maintenance. Advances in spatially-resolved and high-throughput molecular imaging methods such as multiplexed immunofluorescence and spatial transcriptomics (ST) provide exciting new opportunities to augment our fundamental understanding of these processes in health and disease. The large and complex datasets resulting from these techniques, particularly ST, have led to rapid development of innovative machine learning (ML) tools primarily based on deep learning techniques. These ML tools are now increasingly featured in integrated experimental and computational workflows to disentangle signals from noise in complex biological systems. However, it can be difficult to understand and balance the different implicit assumptions and methodologies of a rapidly expanding toolbox of analytical tools in ST. To address this, we summarize major ST analysis goals that ML can help address and current analysis trends. We also describe four major data science concepts and related heuristics that can help guide practitioners in their choices of the right tools for the right biological questions.
Multi-Modal Prototype Learning for Interpretable Multivariable Time Series Classification
Jun 17, 2021
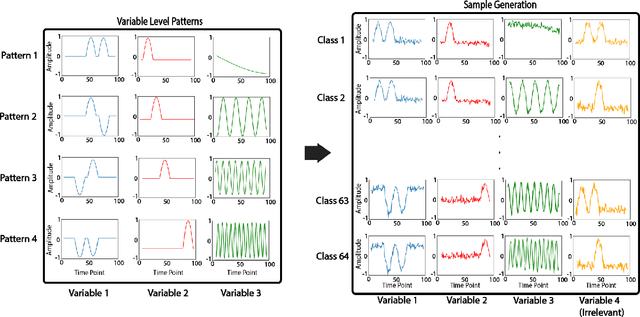
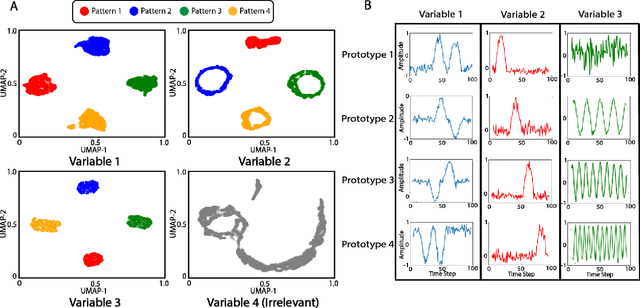
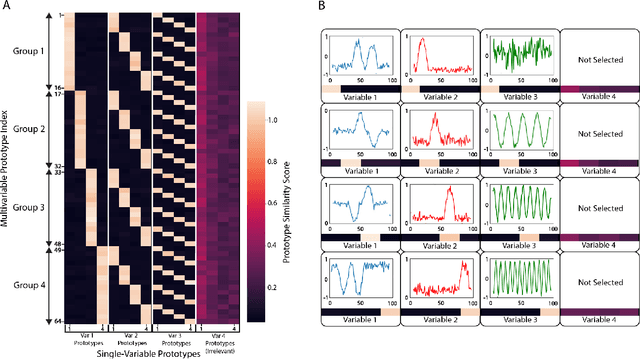
Abstract:Multivariable time series classification problems are increasing in prevalence and complexity in a variety of domains, such as biology and finance. While deep learning methods are an effective tool for these problems, they often lack interpretability. In this work, we propose a novel modular prototype learning framework for multivariable time series classification. In the first stage of our framework, encoders extract features from each variable independently. Prototype layers identify single-variable prototypes in the resulting feature spaces. The next stage of our framework represents the multivariable time series sample points in terms of their similarity to these single-variable prototypes. This results in an inherently interpretable representation of multivariable patterns, on which prototype learning is applied to extract representative examples i.e. multivariable prototypes. Our framework is thus able to explicitly identify both informative patterns in the individual variables, as well as the relationships between the variables. We validate our framework on a simulated dataset with embedded patterns, as well as a real human activity recognition problem. Our framework attains comparable or superior classification performance to existing time series classification methods on these tasks. On the simulated dataset, we find that our model returns interpretations consistent with the embedded patterns. Moreover, the interpretations learned on the activity recognition dataset align with domain knowledge.
Interpretable machine learning: definitions, methods, and applications
Jan 14, 2019
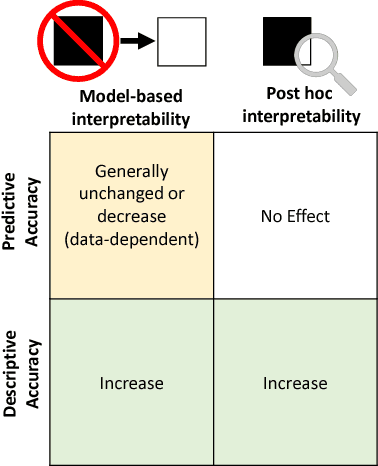
Abstract:Machine-learning models have demonstrated great success in learning complex patterns that enable them to make predictions about unobserved data. In addition to using models for prediction, the ability to interpret what a model has learned is receiving an increasing amount of attention. However, this increased focus has led to considerable confusion about the notion of interpretability. In particular, it is unclear how the wide array of proposed interpretation methods are related, and what common concepts can be used to evaluate them. We aim to address these concerns by defining interpretability in the context of machine learning and introducing the Predictive, Descriptive, Relevant (PDR) framework for discussing interpretations. The PDR framework provides three overarching desiderata for evaluation: predictive accuracy, descriptive accuracy and relevancy, with relevancy judged relative to a human audience. Moreover, to help manage the deluge of interpretation methods, we introduce a categorization of existing techniques into model-based and post-hoc categories, with sub-groups including sparsity, modularity and simulatability. To demonstrate how practitioners can use the PDR framework to evaluate and understand interpretations, we provide numerous real-world examples. These examples highlight the often under-appreciated role played by human audiences in discussions of interpretability. Finally, based on our framework, we discuss limitations of existing methods and directions for future work. We hope that this work will provide a common vocabulary that will make it easier for both practitioners and researchers to discuss and choose from the full range of interpretation methods.
Robust Image Registration via Empirical Mode Decomposition
Nov 12, 2017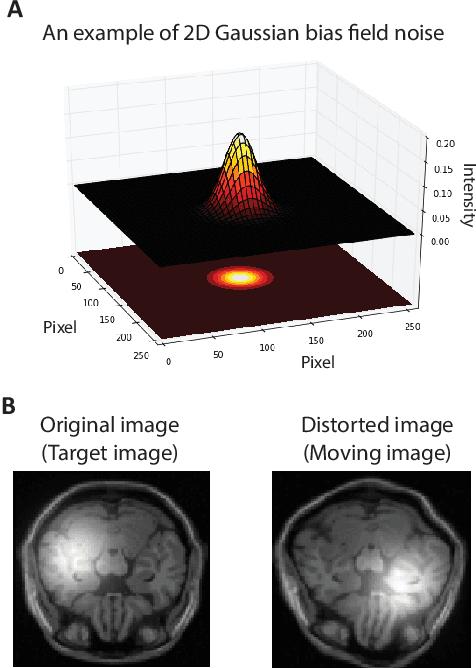
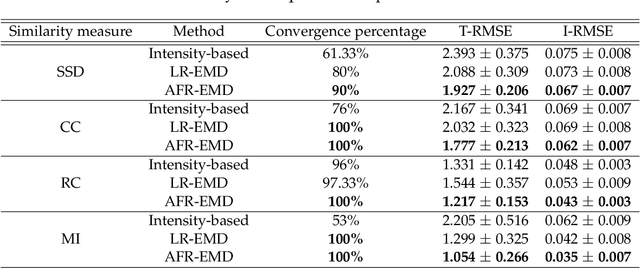
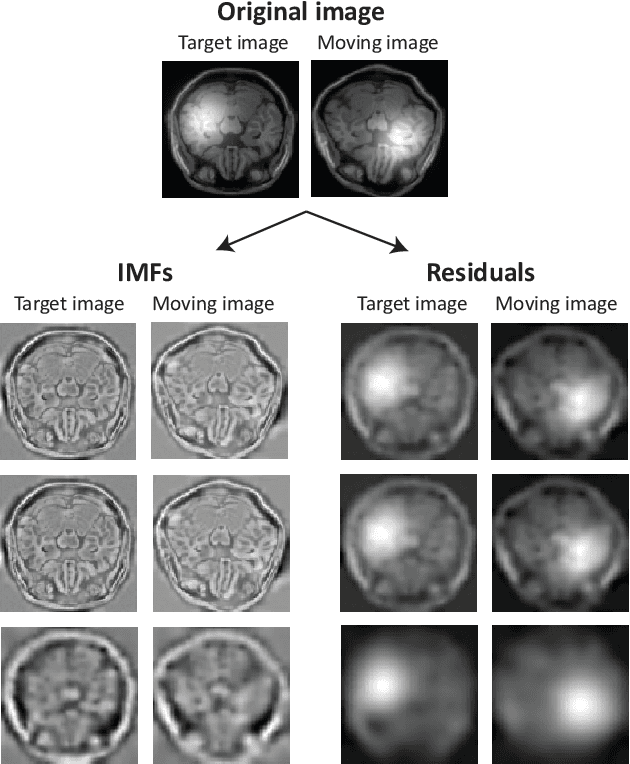
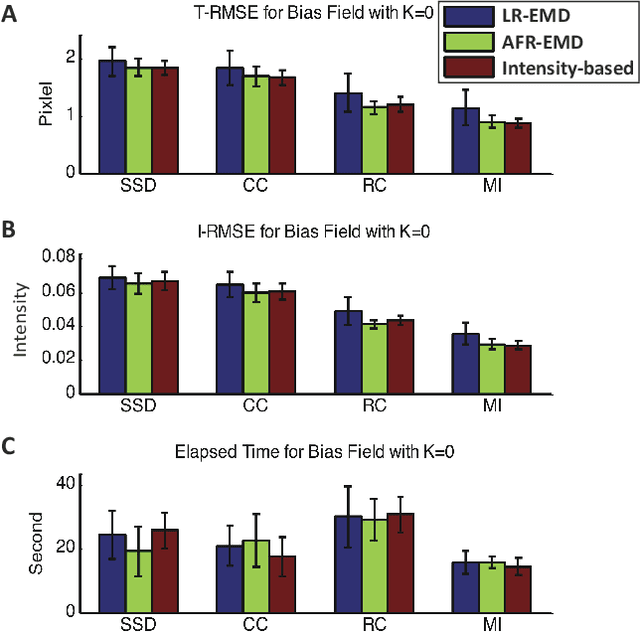
Abstract:Spatially varying intensity noise is a common source of distortion in images. Bias field noise is one example of such distortion that is often present in the magnetic resonance (MR) images. In this paper, we first show that empirical mode decomposition (EMD) can considerably reduce the bias field noise in the MR images. Then, we propose two hierarchical multi-resolution EMD-based algorithms for robust registration of images in the presence of spatially varying noise. One algorithm (LR-EMD) is based on registering EMD feature-maps of both floating and reference images in various resolution levels. In the second algorithm (AFR-EMD), we first extract an average feature-map based on EMD from both floating and reference images. Then, we use a simple hierarchical multi-resolution algorithm based on downsampling to register the average feature-maps. Both algorithms achieve lower error rate and higher convergence percentage compared to the intensity-based hierarchical registration. Specifically, using mutual information as the similarity measure, AFR-EMD achieves 42% lower error rate in intensity and 52% lower error rate in transformation compared to intensity-based hierarchical registration. For LR-EMD, the error rate is 32% lower for the intensity and 41% lower for the transformation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge