Qikui Zhu
Coordinated Transformer with Position \& Sample-aware Central Loss for Anatomical Landmark Detection
May 18, 2023Abstract:Heatmap-based anatomical landmark detection is still facing two unresolved challenges: 1) inability to accurately evaluate the distribution of heatmap; 2) inability to effectively exploit global spatial structure information. To address the computational inability challenge, we propose a novel position-aware and sample-aware central loss. Specifically, our central loss can absorb position information, enabling accurate evaluation of the heatmap distribution. More advanced is that our central loss is sample-aware, which can adaptively distinguish easy and hard samples and make the model more focused on hard samples while solving the challenge of extreme imbalance between landmarks and non-landmarks. To address the challenge of ignoring structure information, a Coordinated Transformer, called CoorTransformer, is proposed, which establishes long-range dependencies under the guidance of landmark coordination information, making the attention more focused on the sparse landmarks while taking advantage of global spatial structure. Furthermore, CoorTransformer can speed up convergence, effectively avoiding the defect that Transformers have difficulty converging in sparse representation learning. Using the advanced CoorTransformer and central loss, we propose a generalized detection model that can handle various scenarios, inherently exploiting the underlying relationship between landmarks and incorporating rich structural knowledge around the target landmarks. We analyzed and evaluated CoorTransformer and central loss on three challenging landmark detection tasks. The experimental results show that our CoorTransformer outperforms state-of-the-art methods, and the central loss significantly improves the performance of the model with p-values< 0.05.
Neural Annotation Refinement: Development of a New 3D Dataset for Adrenal Gland Analysis
Jul 08, 2022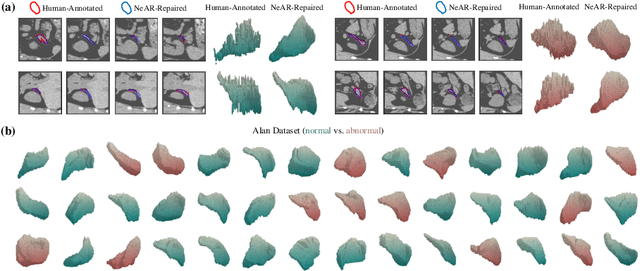
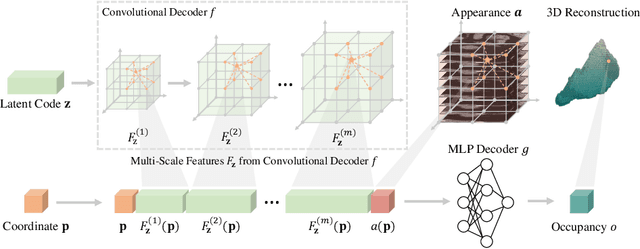
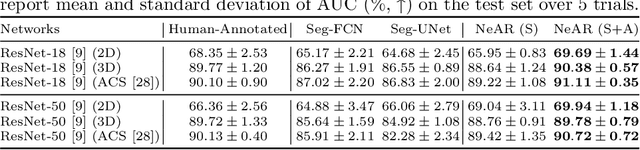

Abstract:The human annotations are imperfect, especially when produced by junior practitioners. Multi-expert consensus is usually regarded as golden standard, while this annotation protocol is too expensive to implement in many real-world projects. In this study, we propose a method to refine human annotation, named Neural Annotation Refinement (NeAR). It is based on a learnable implicit function, which decodes a latent vector into represented shape. By integrating the appearance as an input of implicit functions, the appearance-aware NeAR fixes the annotation artefacts. Our method is demonstrated on the application of adrenal gland analysis. We first show that the NeAR can repair distorted golden standards on a public adrenal gland segmentation dataset. Besides, we develop a new Adrenal gLand ANalysis (ALAN) dataset with the proposed NeAR, where each case consists of a 3D shape of adrenal gland and its diagnosis label (normal vs. abnormal) assigned by experts. We show that models trained on the shapes repaired by the NeAR can diagnose adrenal glands better than the original ones. The ALAN dataset will be open-source, with 1,584 shapes for adrenal gland diagnosis, which serves as a new benchmark for medical shape analysis. Code and dataset are available at https://github.com/M3DV/NeAR.
RRLFSOR: An Efficient Self-Supervised Learning Strategy of Graph Convolutional Networks
Aug 17, 2021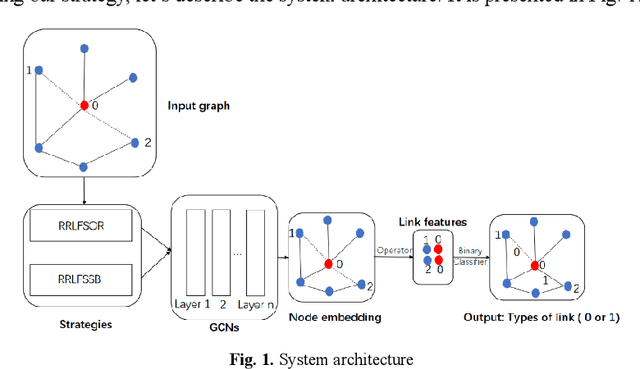
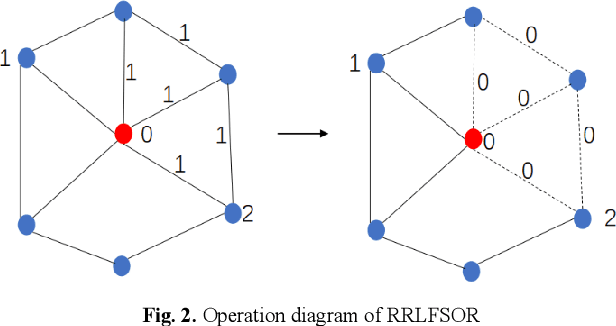
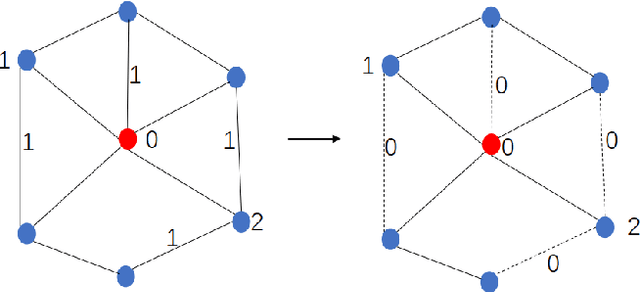

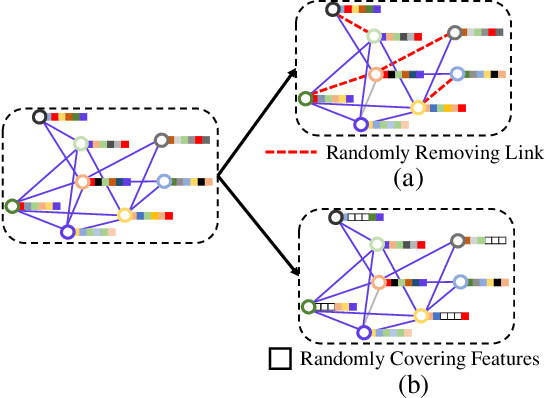
Abstract:To further improve the performance and the self-learning ability of GCNs, in this paper, we propose an efficient self-supervised learning strategy of GCNs, named randomly removed links with a fixed step at one region (RRLFSOR). In addition, we also propose another self-supervised learning strategy of GCNs, named randomly removing links with a fixed step at some blocks (RRLFSSB), to solve the problem that adjacent nodes have no selected step. Experiments on transductive link prediction tasks show that our strategies outperform the baseline models consistently by up to 21.34% in terms of accuracy on three benchmark datasets.
Selective Information Passing for MR/CT Image Segmentation
Oct 10, 2020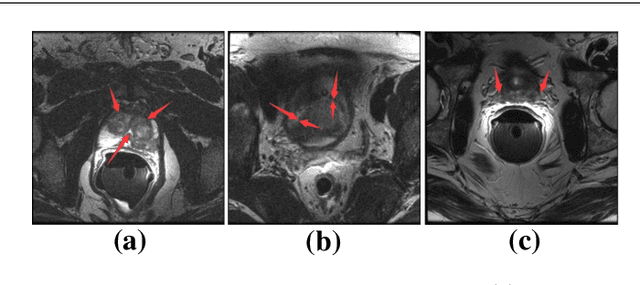
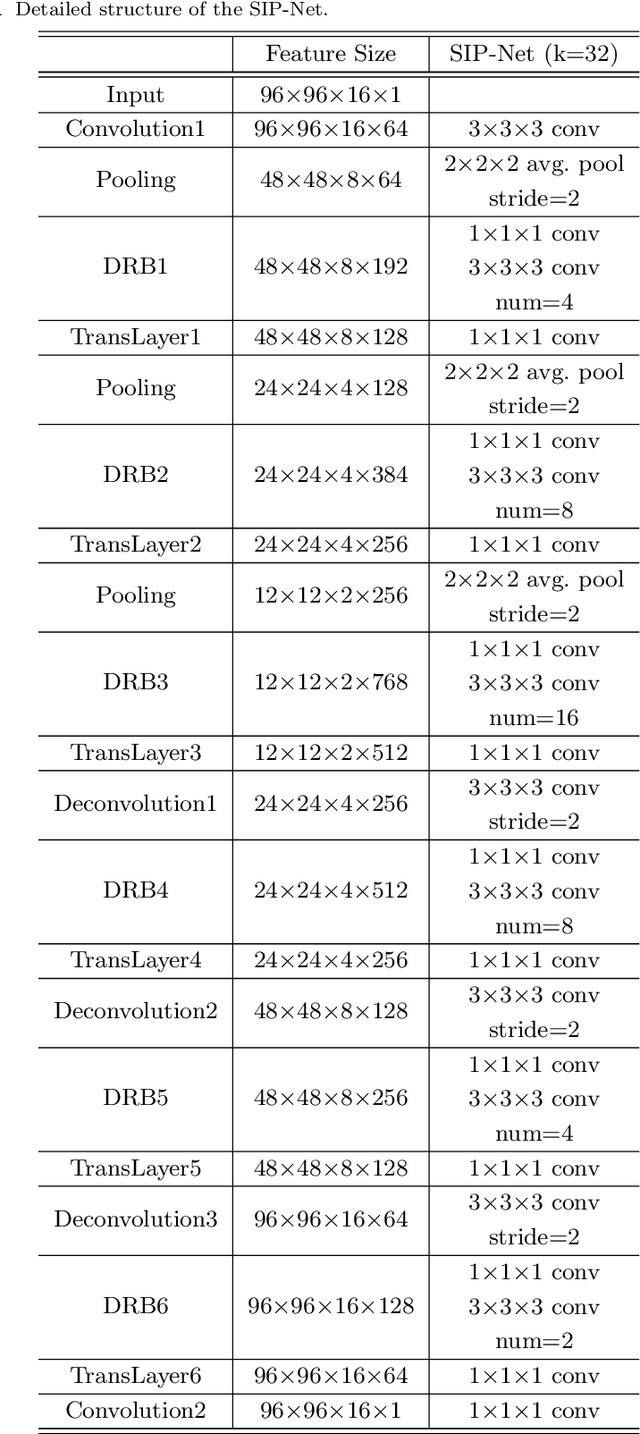
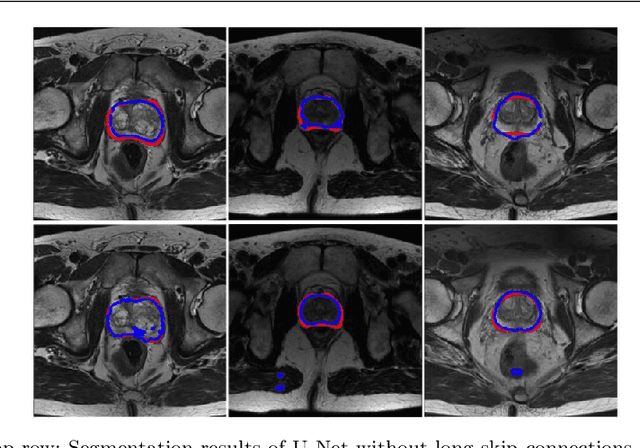
Abstract:Automated medical image segmentation plays an important role in many clinical applications, which however is a very challenging task, due to complex background texture, lack of clear boundary and significant shape and texture variation between images. Many researchers proposed an encoder-decoder architecture with skip connections to combine low-level feature maps from the encoder path with high-level feature maps from the decoder path for automatically segmenting medical images. The skip connections have been shown to be effective in recovering fine-grained details of the target objects and may facilitate the gradient back-propagation. However, not all the feature maps transmitted by those connections contribute positively to the network performance. In this paper, to adaptively select useful information to pass through those skip connections, we propose a novel 3D network with self-supervised function, named selective information passing network (SIP-Net). We evaluate our proposed model on the MICCAI Prostate MR Image Segmentation 2012 Grant Challenge dataset, TCIA Pancreas CT-82 and MICCAI 2017 Liver Tumor Segmentation (LiTS) Challenge dataset. The experimental results across these data sets show that our model achieved improved segmentation results and outperformed other state-of-the-art methods. The source code of this work is available at https://github.com/ahukui/SIPNet.
Self-supervised Training of Graph Convolutional Networks
Jun 03, 2020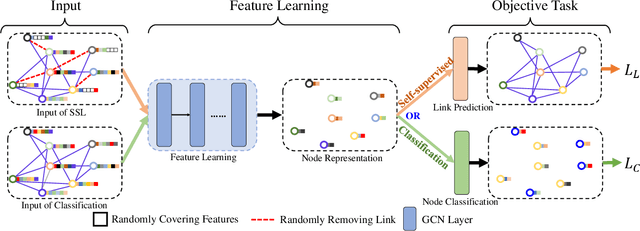
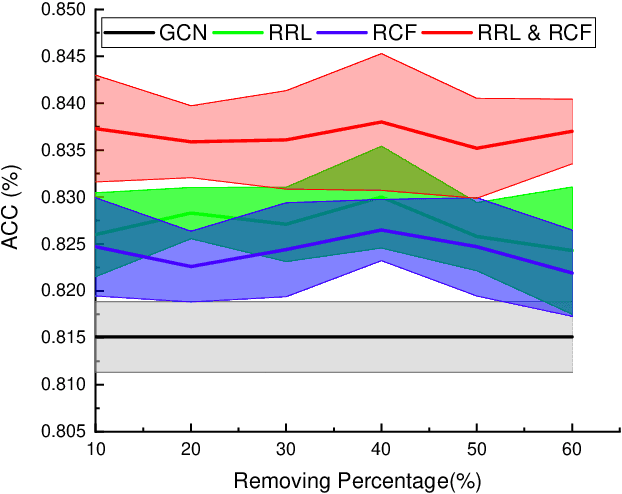
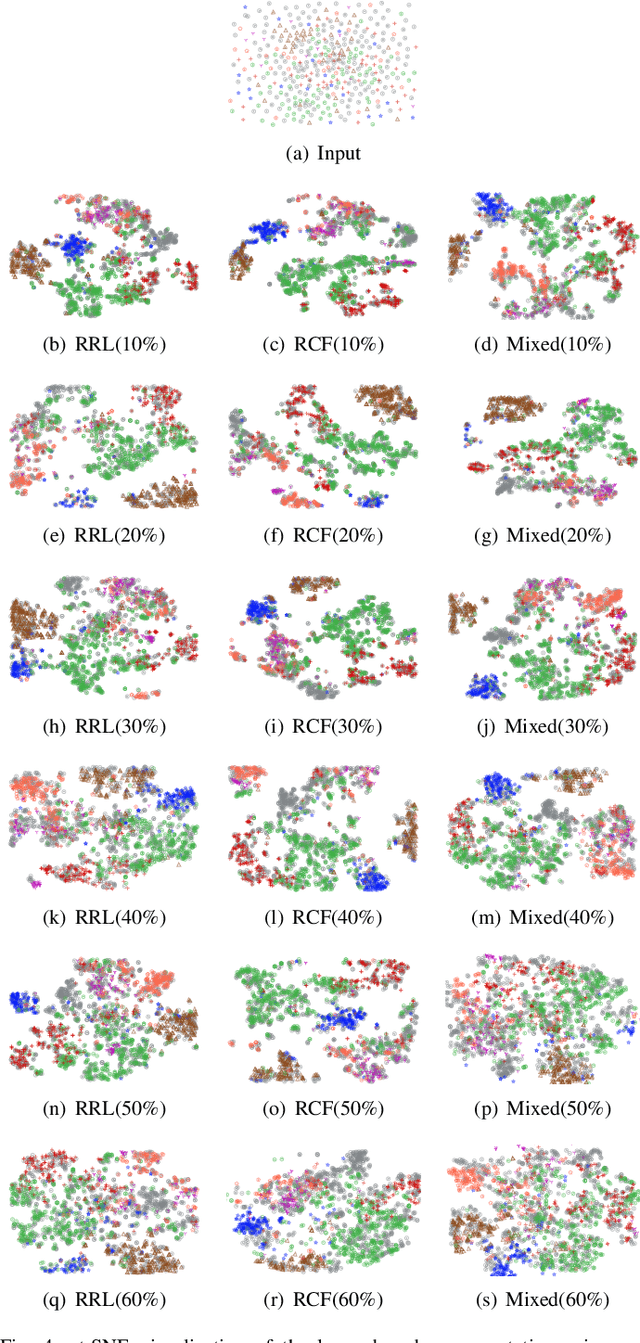
Abstract:Graph Convolutional Networks (GCNs) have been successfully applied to analyze non-grid data, where the classical convolutional neural networks (CNNs) cannot be directly used. One similarity shared by GCNs and CNNs is the requirement of massive amount of labeled data for network training. In addition, GCNs need the adjacency matrix as input to define the relationship between those non-grid data, which leads to all of data including training, validation and test data typically forms only one graph structures data for training. Furthermore, the adjacency matrix is usually pre-defined and stationary, which makes the data augmentation strategies cannot be employed on the constructed graph structures data to augment the amount of training data. To further improve the learning capacity and model performance under the limited training data, in this paper, we propose two types of self-supervised learning strategies to exploit available information from the input graph structure data itself. Our proposed self-supervised learning strategies are examined on two representative GCN models with three public citation network datasets - Citeseer, Cora and Pubmed. The experimental results demonstrate the generalization ability as well as the portability of our proposed strategies, which can significantly improve the performance of GCNs with the power of self-supervised learning in improving feature learning.
OASIS: One-pass aligned Atlas Set for Image Segmentation
Dec 05, 2019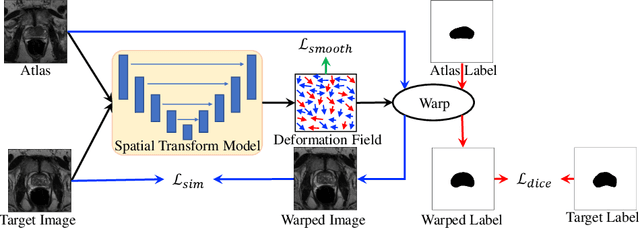
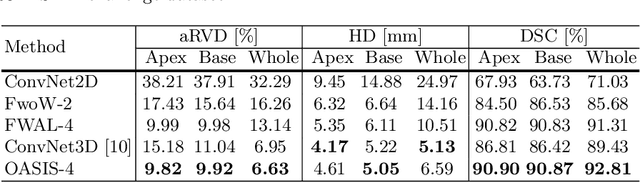
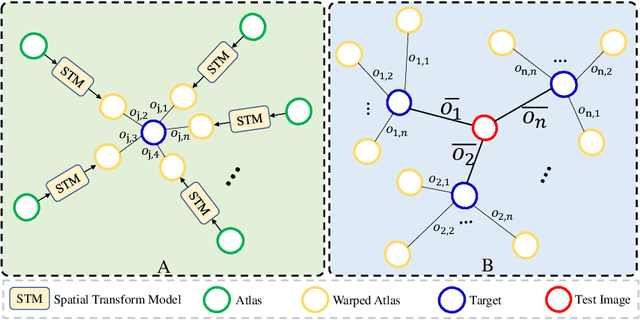
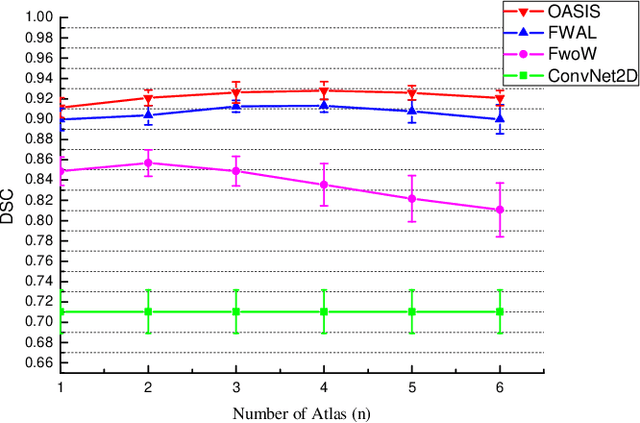
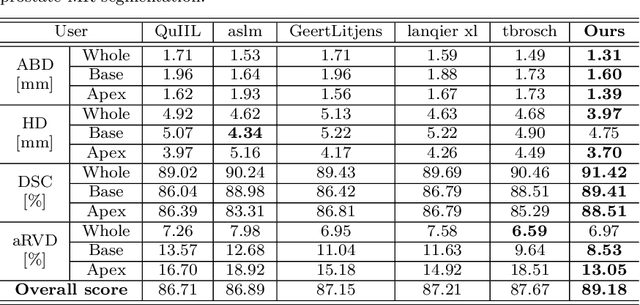
Abstract:Medical image segmentation is a fundamental task in medical image analysis. Despite that deep convolutional neural networks have gained stellar performance in this challenging task, they typically rely on large labeled datasets, which have limited their extension to customized applications. By revisiting the superiority of atlas based segmentation methods, we present a new framework of One-pass aligned Atlas Set for Images Segmentation (OASIS). To address the problem of time-consuming iterative image registration used for atlas warping, the proposed method takes advantage of the power of deep learning to achieve one-pass image registration. In addition, by applying label constraint, OASIS also makes the registration process to be focused on the regions to be segmented for improving the performance of segmentation. Furthermore, instead of using image based similarity for label fusion, which can be distracted by the large background areas, we propose a novel strategy to compute the label similarity based weights for label fusion. Our experimental results on the challenging task of prostate MR image segmentation demonstrate that OASIS is able to significantly increase the segmentation performance compared to other state-of-the-art methods.
Multi-hop Convolutions on Weighted Graphs
Nov 12, 2019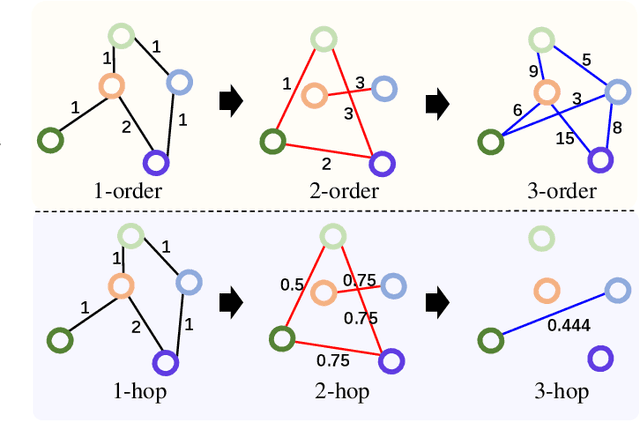
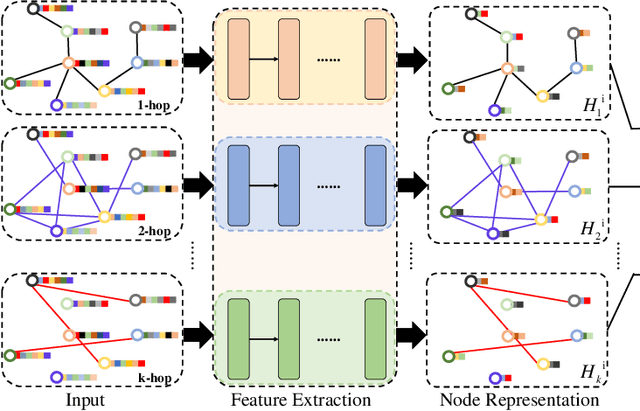
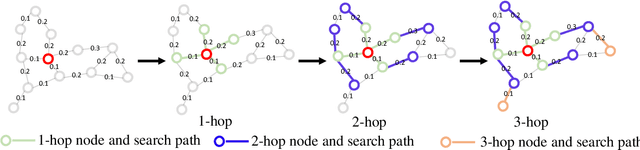
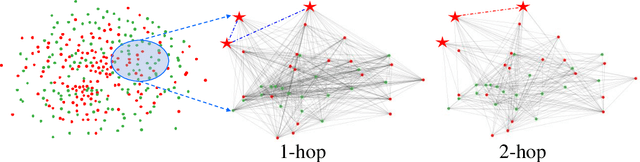
Abstract:Graph Convolutional Networks (GCNs) have made significant advances in semi-supervised learning, especially for classification tasks. However, existing GCN based methods have two main drawbacks. First, to increase the receptive field and improve the representation capability of GCNs, larger kernels or deeper network architectures are used, which greatly increases the computational complexity and the number of parameters. Second, methods working on higher order graphs computed directly from adjacency matrices may alter the relationship between graph nodes, particularly for weighted graphs. In addition, the direct construction of higher-order graphs introduces redundant information, which may result in lower network performance. To address the above weaknesses, in this paper, we propose a new method of multi-hop convolutional network on weighted graphs. The proposed method consists of multiple convolutional branches, where each branch extracts node representation from a $k$-hop graph with small kernels. Such design systematically aggregates multi-scale contextual information without adding redundant information. Furthermore, to efficiently combine the extracted information from the multi-hop branches, an adaptive weight computation (AWC) layer is proposed. We demonstrate the superiority of our MultiHop in six publicly available datasets, including three citation network datasets and three medical image datasets. The experimental results show that our proposed MultiHop method achieves the highest classification accuracy and outperforms the state-of-the-art methods. The source code of this work have been released on GitHub (https://github.com/ahukui/Multi-hop-Convolutions-on-Weighted-Graphs).
Boundary-weighted Domain Adaptive Neural Network for Prostate MR Image Segmentation
Feb 21, 2019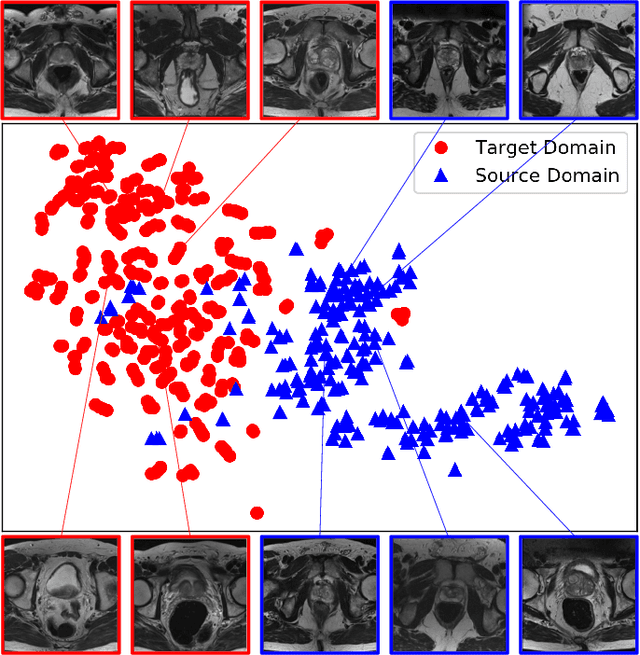
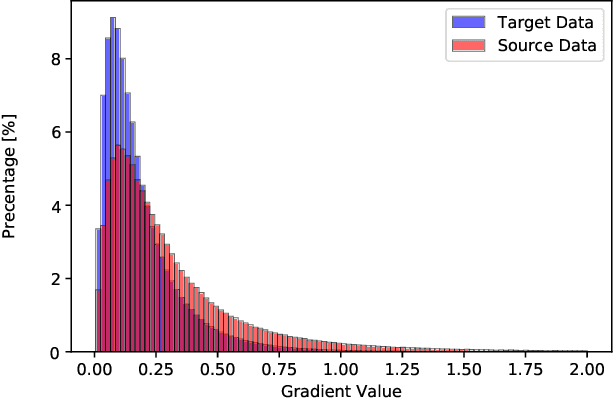
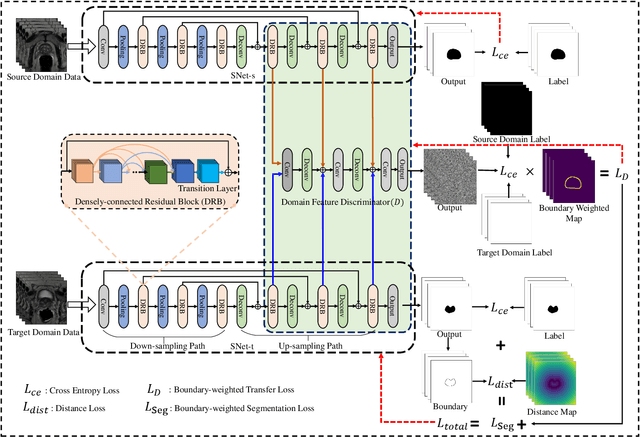
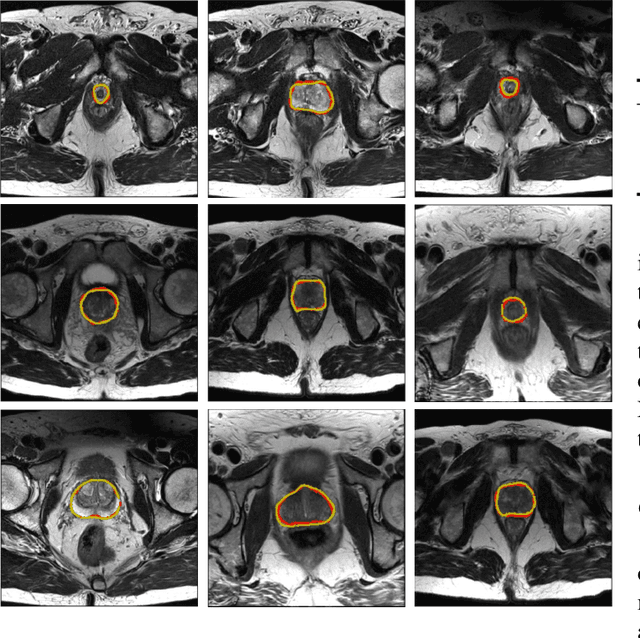
Abstract:Accurate segmentation of the prostate from magnetic resonance (MR) images provides useful information for prostate cancer diagnosis and treatment. However, automated prostate segmentation from 3D MR images still faces several challenges. For instance, a lack of clear edge between the prostate and other anatomical structures makes it challenging to accurately extract the boundaries. The complex background texture and large variation in size, shape and intensity distribution of the prostate itself make segmentation even further complicated. With deep learning, especially convolutional neural networks (CNNs), emerging as commonly used methods for medical image segmentation, the difficulty in obtaining large number of annotated medical images for training CNNs has become much more pronounced that ever before. Since large-scale dataset is one of the critical components for the success of deep learning, lack of sufficient training data makes it difficult to fully train complex CNNs. To tackle the above challenges, in this paper, we propose a boundary-weighted domain adaptive neural network (BOWDA-Net). To make the network more sensitive to the boundaries during segmentation, a boundary-weighted segmentation loss (BWL) is proposed. Furthermore, an advanced boundary-weighted transfer leaning approach is introduced to address the problem of small medical imaging datasets. We evaluate our proposed model on the publicly available MICCAI 2012 Prostate MR Image Segmentation (PROMISE12) challenge dataset. Our experimental results demonstrate that the proposed model is more sensitive to boundary information and outperformed other state-of-the-art methods.
Deeply-Supervised CNN for Prostate Segmentation
Mar 28, 2017
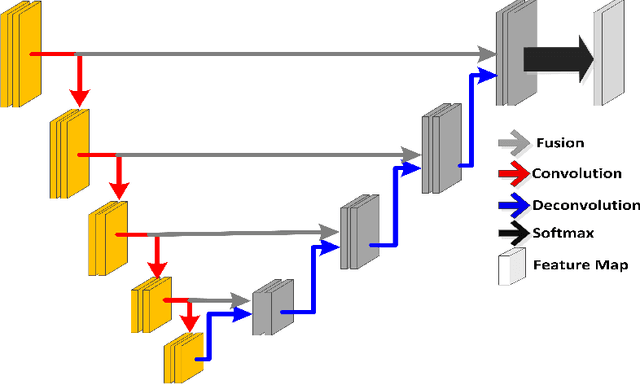
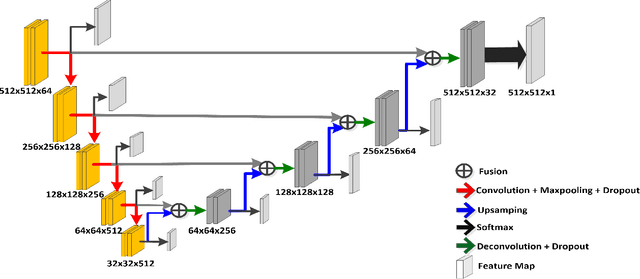
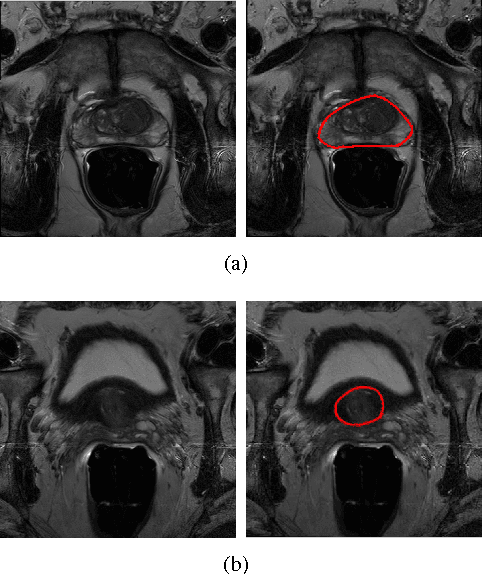
Abstract:Prostate segmentation from Magnetic Resonance (MR) images plays an important role in image guided interven- tion. However, the lack of clear boundary specifically at the apex and base, and huge variation of shape and texture between the images from different patients make the task very challenging. To overcome these problems, in this paper, we propose a deeply supervised convolutional neural network (CNN) utilizing the convolutional information to accurately segment the prostate from MR images. The proposed model can effectively detect the prostate region with additional deeply supervised layers compared with other approaches. Since some information will be abandoned after convolution, it is necessary to pass the features extracted from early stages to later stages. The experimental results show that significant segmentation accuracy improvement has been achieved by our proposed method compared to other reported approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge