Qichao Zhou
Indescribable Multi-modal Spatial Evaluator
Mar 02, 2023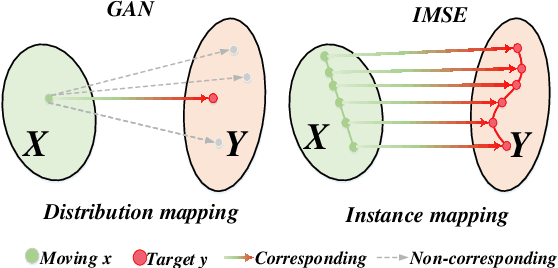
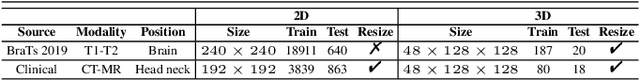
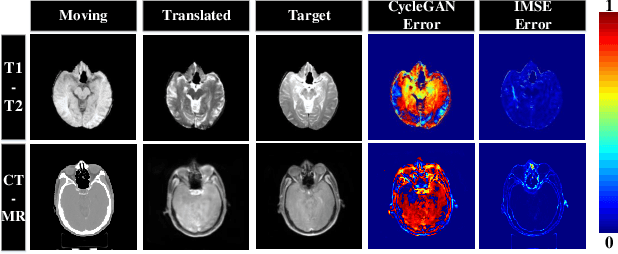
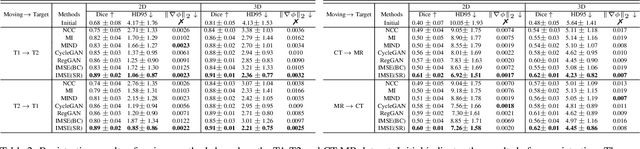
Abstract:Multi-modal image registration spatially aligns two images with different distributions. One of its major challenges is that images acquired from different imaging machines have different imaging distributions, making it difficult to focus only on the spatial aspect of the images and ignore differences in distributions. In this study, we developed a self-supervised approach, Indescribable Multi-model Spatial Evaluator (IMSE), to address multi-modal image registration. IMSE creates an accurate multi-modal spatial evaluator to measure spatial differences between two images, and then optimizes registration by minimizing the error predicted of the evaluator. To optimize IMSE performance, we also proposed a new style enhancement method called Shuffle Remap which randomizes the image distribution into multiple segments, and then randomly disorders and remaps these segments, so that the distribution of the original image is changed. Shuffle Remap can help IMSE to predict the difference in spatial location from unseen target distributions. Our results show that IMSE outperformed the existing methods for registration using T1-T2 and CT-MRI datasets. IMSE also can be easily integrated into the traditional registration process, and can provide a convenient way to evaluate and visualize registration results. IMSE also has the potential to be used as a new paradigm for image-to-image translation. Our code is available at https://github.com/Kid-Liet/IMSE.
XBound-Former: Toward Cross-scale Boundary Modeling in Transformers
Jun 02, 2022



Abstract:Skin lesion segmentation from dermoscopy images is of great significance in the quantitative analysis of skin cancers, which is yet challenging even for dermatologists due to the inherent issues, i.e., considerable size, shape and color variation, and ambiguous boundaries. Recent vision transformers have shown promising performance in handling the variation through global context modeling. Still, they have not thoroughly solved the problem of ambiguous boundaries as they ignore the complementary usage of the boundary knowledge and global contexts. In this paper, we propose a novel cross-scale boundary-aware transformer, \textbf{XBound-Former}, to simultaneously address the variation and boundary problems of skin lesion segmentation. XBound-Former is a purely attention-based network and catches boundary knowledge via three specially designed learners. We evaluate the model on two skin lesion datasets, ISIC-2016\&PH$^2$ and ISIC-2018, where our model consistently outperforms other convolution- and transformer-based models, especially on the boundary-wise metrics. We extensively verify the generalization ability of polyp lesion segmentation that has similar characteristics, and our model can also yield significant improvement compared to the latest models.
Separated Contrastive Learning for Organ-at-Risk and Gross-Tumor-Volume Segmentation with Limited Annotation
Dec 06, 2021



Abstract:Automatic delineation of organ-at-risk (OAR) and gross-tumor-volume (GTV) is of great significance for radiotherapy planning. However, it is a challenging task to learn powerful representations for accurate delineation under limited pixel (voxel)-wise annotations. Contrastive learning at pixel-level can alleviate the dependency on annotations by learning dense representations from unlabeled data. Recent studies in this direction design various contrastive losses on the feature maps, to yield discriminative features for each pixel in the map. However, pixels in the same map inevitably share semantics to be closer than they actually are, which may affect the discrimination of pixels in the same map and lead to the unfair comparison to pixels in other maps. To address these issues, we propose a separated region-level contrastive learning scheme, namely SepaReg, the core of which is to separate each image into regions and encode each region separately. Specifically, SepaReg comprises two components: a structure-aware image separation (SIS) module and an intra- and inter-organ distillation (IID) module. The SIS is proposed to operate on the image set to rebuild a region set under the guidance of structural information. The inter-organ representation will be learned from this set via typical contrastive losses cross regions. On the other hand, the IID is proposed to tackle the quantity imbalance in the region set as tiny organs may produce fewer regions, by exploiting intra-organ representations. We conducted extensive experiments to evaluate the proposed model on a public dataset and two private datasets. The experimental results demonstrate the effectiveness of the proposed model, consistently achieving better performance than state-of-the-art approaches. Code is available at https://github.com/jcwang123/Separate_CL.
Breaking the Dilemma of Medical Image-to-image Translation
Oct 13, 2021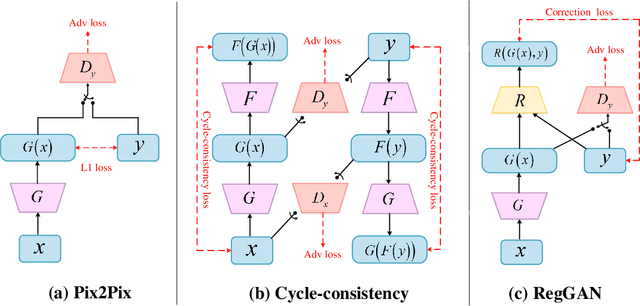
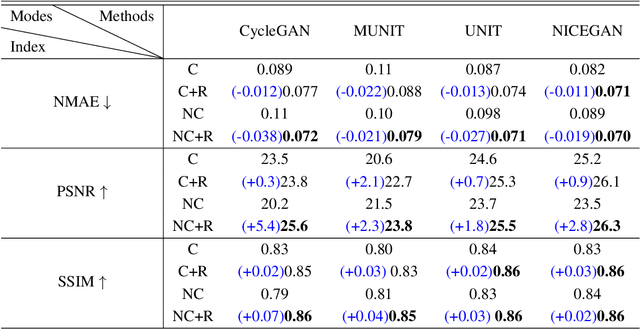
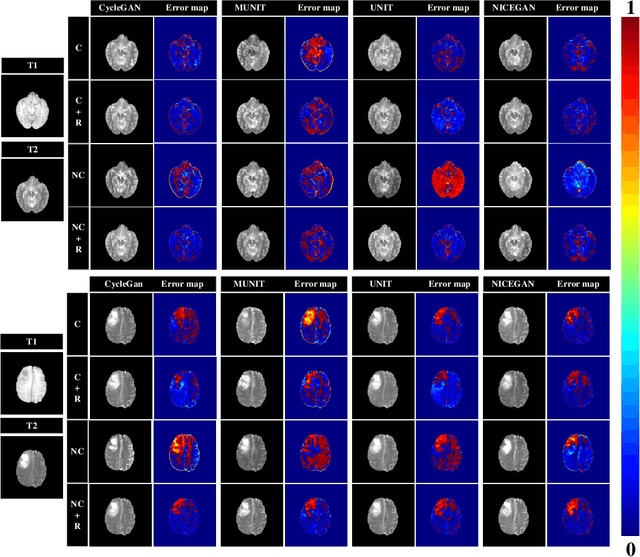
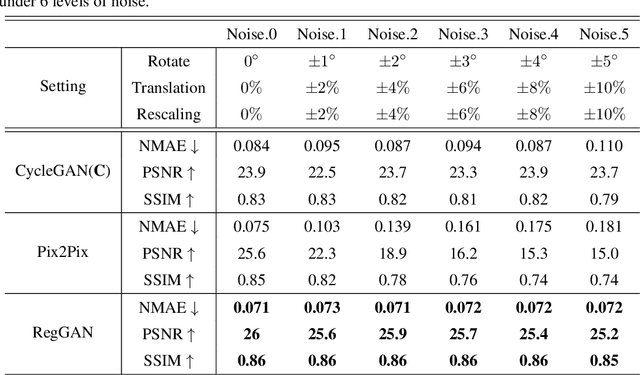
Abstract:Supervised Pix2Pix and unsupervised Cycle-consistency are two modes that dominate the field of medical image-to-image translation. However, neither modes are ideal. The Pix2Pix mode has excellent performance. But it requires paired and well pixel-wise aligned images, which may not always be achievable due to respiratory motion or anatomy change between times that paired images are acquired. The Cycle-consistency mode is less stringent with training data and works well on unpaired or misaligned images. But its performance may not be optimal. In order to break the dilemma of the existing modes, we propose a new unsupervised mode called RegGAN for medical image-to-image translation. It is based on the theory of "loss-correction". In RegGAN, the misaligned target images are considered as noisy labels and the generator is trained with an additional registration network to fit the misaligned noise distribution adaptively. The goal is to search for the common optimal solution to both image-to-image translation and registration tasks. We incorporated RegGAN into a few state-of-the-art image-to-image translation methods and demonstrated that RegGAN could be easily combined with these methods to improve their performances. Such as a simple CycleGAN in our mode surpasses latest NICEGAN even though using less network parameters. Based on our results, RegGAN outperformed both Pix2Pix on aligned data and Cycle-consistency on misaligned or unpaired data. RegGAN is insensitive to noises which makes it a better choice for a wide range of scenarios, especially for medical image-to-image translation tasks in which well pixel-wise aligned data are not available
Boundary-aware Transformers for Skin Lesion Segmentation
Oct 08, 2021



Abstract:Skin lesion segmentation from dermoscopy images is of great importance for improving the quantitative analysis of skin cancer. However, the automatic segmentation of melanoma is a very challenging task owing to the large variation of melanoma and ambiguous boundaries of lesion areas. While convolutional neutral networks (CNNs) have achieved remarkable progress in this task, most of existing solutions are still incapable of effectively capturing global dependencies to counteract the inductive bias caused by limited receptive fields. Recently, transformers have been proposed as a promising tool for global context modeling by employing a powerful global attention mechanism, but one of their main shortcomings when applied to segmentation tasks is that they cannot effectively extract sufficient local details to tackle ambiguous boundaries. We propose a novel boundary-aware transformer (BAT) to comprehensively address the challenges of automatic skin lesion segmentation. Specifically, we integrate a new boundary-wise attention gate (BAG) into transformers to enable the whole network to not only effectively model global long-range dependencies via transformers but also, simultaneously, capture more local details by making full use of boundary-wise prior knowledge. Particularly, the auxiliary supervision of BAG is capable of assisting transformers to learn position embedding as it provides much spatial information. We conducted extensive experiments to evaluate the proposed BAT and experiments corroborate its effectiveness, consistently outperforming state-of-the-art methods in two famous datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge