X. Sharon Qi
Indescribable Multi-modal Spatial Evaluator
Mar 02, 2023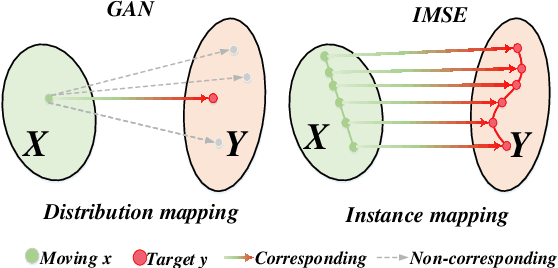
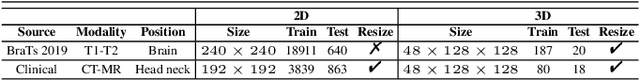
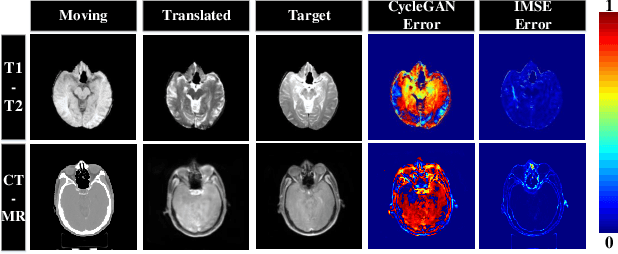
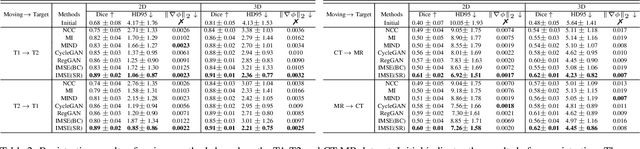
Abstract:Multi-modal image registration spatially aligns two images with different distributions. One of its major challenges is that images acquired from different imaging machines have different imaging distributions, making it difficult to focus only on the spatial aspect of the images and ignore differences in distributions. In this study, we developed a self-supervised approach, Indescribable Multi-model Spatial Evaluator (IMSE), to address multi-modal image registration. IMSE creates an accurate multi-modal spatial evaluator to measure spatial differences between two images, and then optimizes registration by minimizing the error predicted of the evaluator. To optimize IMSE performance, we also proposed a new style enhancement method called Shuffle Remap which randomizes the image distribution into multiple segments, and then randomly disorders and remaps these segments, so that the distribution of the original image is changed. Shuffle Remap can help IMSE to predict the difference in spatial location from unseen target distributions. Our results show that IMSE outperformed the existing methods for registration using T1-T2 and CT-MRI datasets. IMSE also can be easily integrated into the traditional registration process, and can provide a convenient way to evaluate and visualize registration results. IMSE also has the potential to be used as a new paradigm for image-to-image translation. Our code is available at https://github.com/Kid-Liet/IMSE.
Deep Learning-based Radiomic Features for Improving Neoadjuvant Chemoradiation Response Prediction in Locally Advanced Rectal Cancer
Sep 09, 2019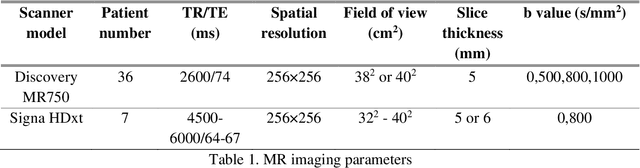
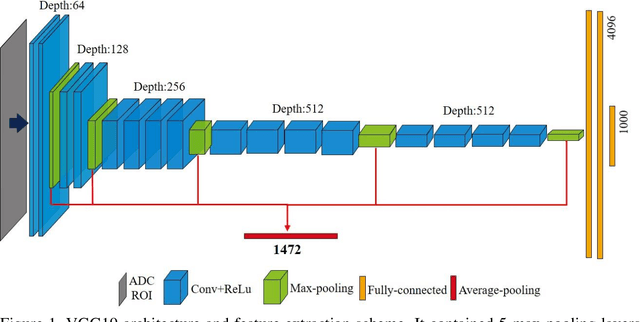
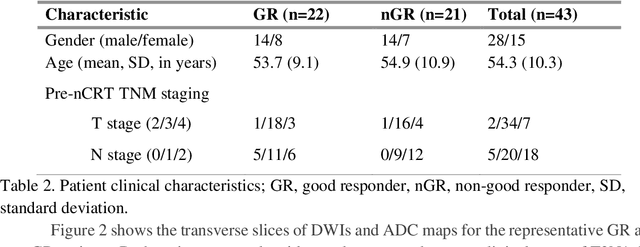
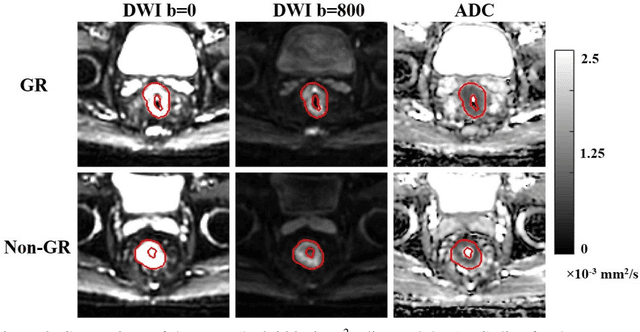
Abstract:Radiomic features achieve promising results in cancer diagnosis, treatment response prediction, and survival prediction. Our goal is to compare the handcrafted (explicitly designed) and deep learning (DL)-based radiomic features extracted from pre-treatment diffusion-weighted magnetic resonance images (DWIs) for predicting neoadjuvant chemoradiation treatment (nCRT) response in patients with locally advanced rectal cancer (LARC). 43 patients receiving nCRT were included. All patients underwent DWIs before nCRT and total mesorectal excision surgery 6-12 weeks after completion of nCRT. Gross tumor volume (GTV) contours were drawn by an experienced radiation oncologist on DWIs. The patient-cohort was split into the responder group (n=22) and the non-responder group (n=21) based on the post-nCRT response assessed by postoperative pathology, MRI or colonoscopy. Handcrafted and DL-based features were extracted from the apparent diffusion coefficient (ADC) map of the DWI using conventional computer-aided diagnosis methods and a pre-trained convolution neural network, respectively. Least absolute shrinkage and selection operator (LASSO)-logistic regression models were constructed using extracted features for predicting treatment response. The model performance was evaluated with repeated 20 times stratified 4-fold cross-validation using receiver operating characteristic (ROC) curves and compared using the corrected resampled t-test. The model built with handcrafted features achieved the mean area under the ROC curve (AUC) of 0.64, while the one built with DL-based features yielded the mean AUC of 0.73. The corrected resampled t-test on AUC showed P-value < 0.05. DL-based features extracted from pre-treatment DWIs achieved significantly better classification performance compared with handcrafted features for predicting nCRT response in patients with LARC.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge