John Lewis
Frontier Shepherding: A Bio-Mimetic Multi-robot Framework for Large-Scale Exploration
Sep 17, 2024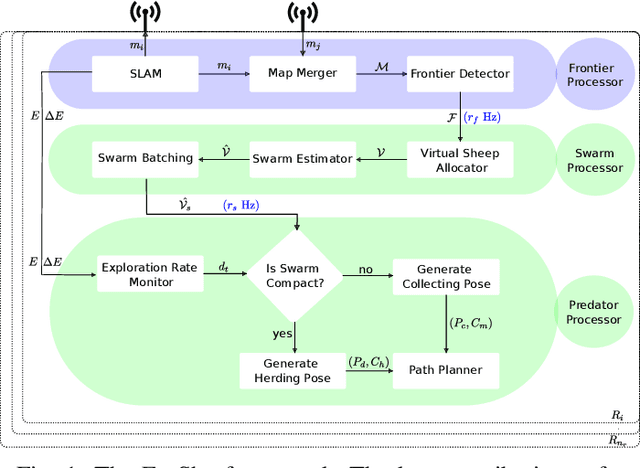
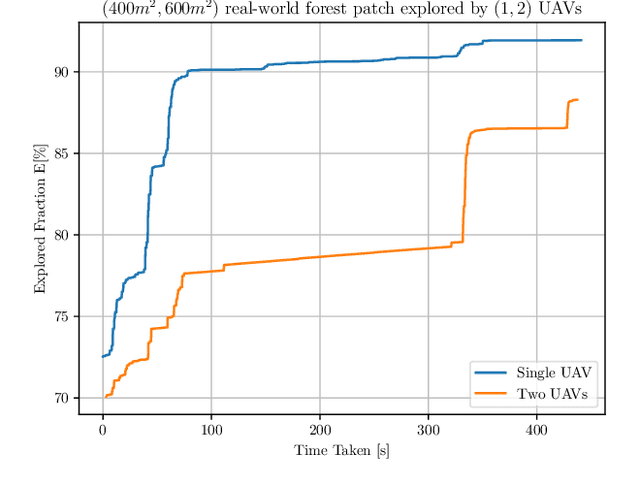
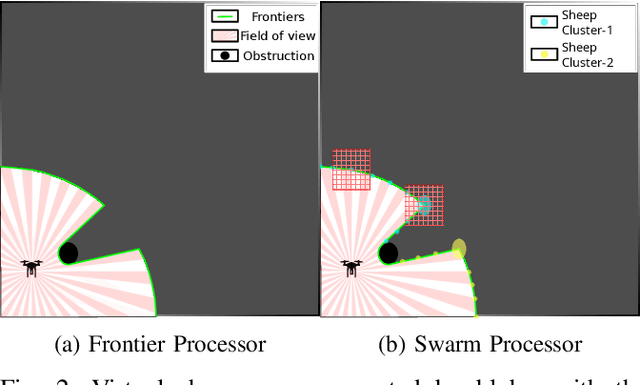
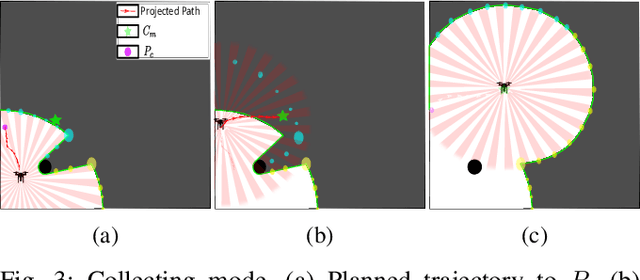
Abstract:Efficient exploration of large-scale environments remains a critical challenge in robotics, with applications ranging from environmental monitoring to search and rescue operations. This article proposes a bio-mimetic multi-robot framework, \textit{Frontier Shepherding (FroShe)}, for large-scale exploration. The presented bio-inspired framework heuristically models frontier exploration similar to the shepherding behavior of herding dogs. This is achieved by modeling frontiers as a sheep swarm reacting to robots modeled as shepherding dogs. The framework is robust across varying environment sizes and obstacle densities and can be easily deployed across multiple agents. Simulation results showcase that the proposed method consistently performed irrespective of the simulated environment's varying sizes and obstacle densities. With the increase in the number of agents, the proposed method outperforms other state-of-the-art exploration methods, with an average improvement of $20\%$ with the next-best approach(for $3$ UAVs). The proposed technique was implemented and tested in a single and dual drone scenario in a real-world forest-like environment.
LipSync3D: Data-Efficient Learning of Personalized 3D Talking Faces from Video using Pose and Lighting Normalization
Jun 08, 2021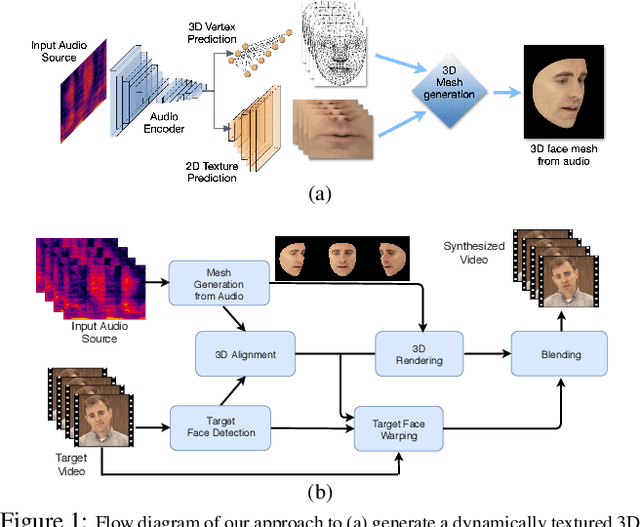
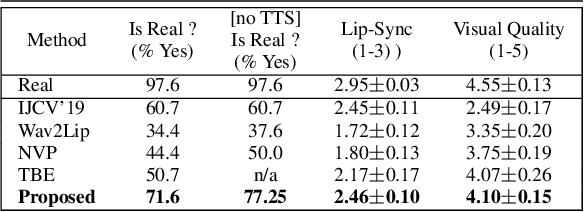
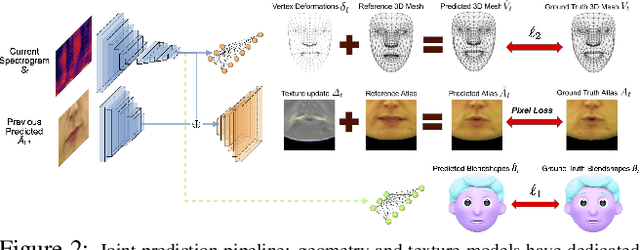

Abstract:In this paper, we present a video-based learning framework for animating personalized 3D talking faces from audio. We introduce two training-time data normalizations that significantly improve data sample efficiency. First, we isolate and represent faces in a normalized space that decouples 3D geometry, head pose, and texture. This decomposes the prediction problem into regressions over the 3D face shape and the corresponding 2D texture atlas. Second, we leverage facial symmetry and approximate albedo constancy of skin to isolate and remove spatio-temporal lighting variations. Together, these normalizations allow simple networks to generate high fidelity lip-sync videos under novel ambient illumination while training with just a single speaker-specific video. Further, to stabilize temporal dynamics, we introduce an auto-regressive approach that conditions the model on its previous visual state. Human ratings and objective metrics demonstrate that our method outperforms contemporary state-of-the-art audio-driven video reenactment benchmarks in terms of realism, lip-sync and visual quality scores. We illustrate several applications enabled by our framework.
Deep Learning-based Radiomic Features for Improving Neoadjuvant Chemoradiation Response Prediction in Locally Advanced Rectal Cancer
Sep 09, 2019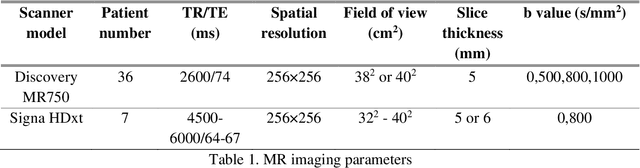
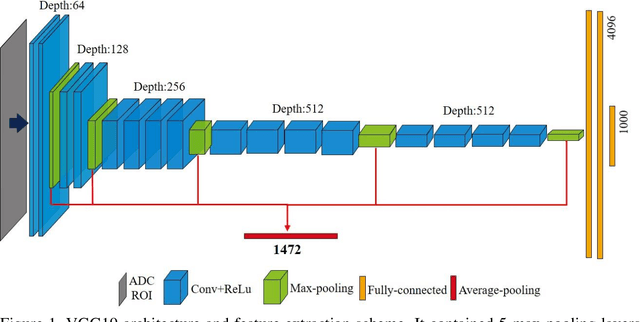
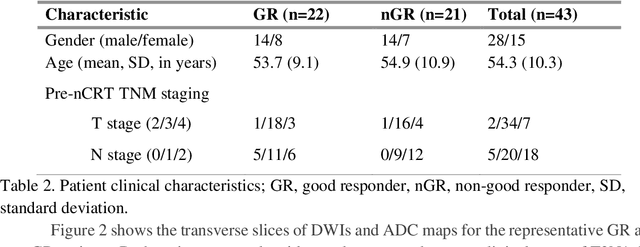
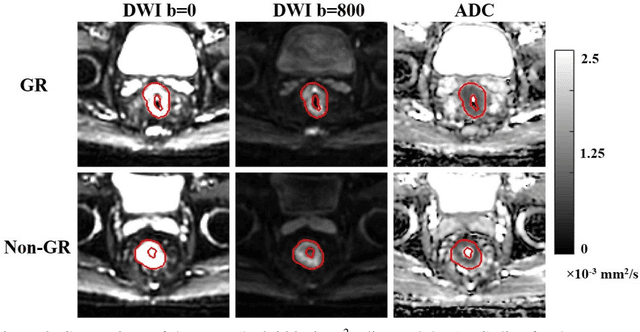
Abstract:Radiomic features achieve promising results in cancer diagnosis, treatment response prediction, and survival prediction. Our goal is to compare the handcrafted (explicitly designed) and deep learning (DL)-based radiomic features extracted from pre-treatment diffusion-weighted magnetic resonance images (DWIs) for predicting neoadjuvant chemoradiation treatment (nCRT) response in patients with locally advanced rectal cancer (LARC). 43 patients receiving nCRT were included. All patients underwent DWIs before nCRT and total mesorectal excision surgery 6-12 weeks after completion of nCRT. Gross tumor volume (GTV) contours were drawn by an experienced radiation oncologist on DWIs. The patient-cohort was split into the responder group (n=22) and the non-responder group (n=21) based on the post-nCRT response assessed by postoperative pathology, MRI or colonoscopy. Handcrafted and DL-based features were extracted from the apparent diffusion coefficient (ADC) map of the DWI using conventional computer-aided diagnosis methods and a pre-trained convolution neural network, respectively. Least absolute shrinkage and selection operator (LASSO)-logistic regression models were constructed using extracted features for predicting treatment response. The model performance was evaluated with repeated 20 times stratified 4-fold cross-validation using receiver operating characteristic (ROC) curves and compared using the corrected resampled t-test. The model built with handcrafted features achieved the mean area under the ROC curve (AUC) of 0.64, while the one built with DL-based features yielded the mean AUC of 0.73. The corrected resampled t-test on AUC showed P-value < 0.05. DL-based features extracted from pre-treatment DWIs achieved significantly better classification performance compared with handcrafted features for predicting nCRT response in patients with LARC.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge