Pinaki Sarder
Department of Pathology & Anatomical Sciences, University at Buffalo, the State University of New York, Buffalo, New York, Department of Biostatistics, University at Buffalo, the State University of New York, Buffalo, New York, Department of Biomedical Engineering, University at Buffalo, the State University of New York, Buffalo, New York
CASC-AI: Consensus-aware Self-corrective AI Agents for Noise Cell Segmentation
Feb 11, 2025Abstract:Multi-class cell segmentation in high-resolution gigapixel whole slide images (WSI) is crucial for various clinical applications. However, training such models typically requires labor-intensive, pixel-wise annotations by domain experts. Recent efforts have democratized this process by involving lay annotators without medical expertise. However, conventional non-agent-based approaches struggle to handle annotation noise adaptively, as they lack mechanisms to mitigate false positives (FP) and false negatives (FN) at both the image-feature and pixel levels. In this paper, we propose a consensus-aware self-corrective AI agent that leverages the Consensus Matrix to guide its learning process. The Consensus Matrix defines regions where both the AI and annotators agree on cell and non-cell annotations, which are prioritized with stronger supervision. Conversely, areas of disagreement are adaptively weighted based on their feature similarity to high-confidence agreement regions, with more similar regions receiving greater attention. Additionally, contrastive learning is employed to separate features of noisy regions from those of reliable agreement regions by maximizing their dissimilarity. This paradigm enables the AI to iteratively refine noisy labels, enhancing its robustness. Validated on one real-world lay-annotated cell dataset and two simulated noisy datasets, our method demonstrates improved segmentation performance, effectively correcting FP and FN errors and showcasing its potential for training robust models on noisy datasets. The official implementation and cell annotations are publicly available at https://github.com/ddrrnn123/CASC-AI.
Histo-fetch -- On-the-fly processing of gigapixel whole slide images simplifies and speeds neural network training
Mar 01, 2021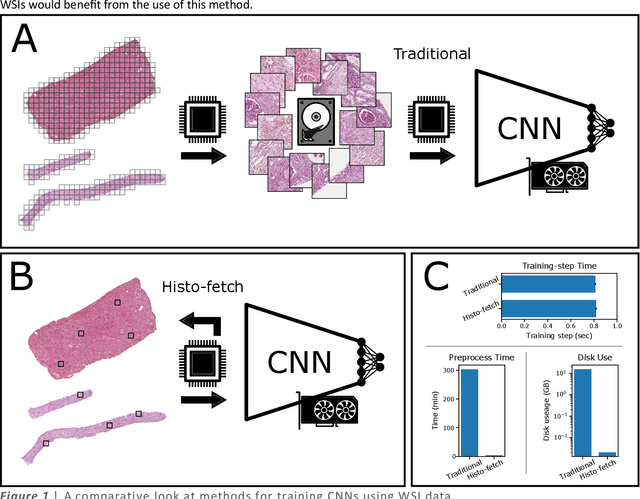

Abstract:We created a custom pipeline (histo-fetch) to efficiently extract random patches and labels from pathology whole slide images (WSIs) for input to a neural network on-the-fly. We prefetch these patches as needed during network training, avoiding the need for WSI preparation such as chopping/tiling. We demonstrate the utility of this pipeline to perform artificial stain transfer and image generation using the popular networks CycleGAN and ProGAN, respectively.
A tool for user friendly, cloud based, whole slide image segmentation
Jan 18, 2021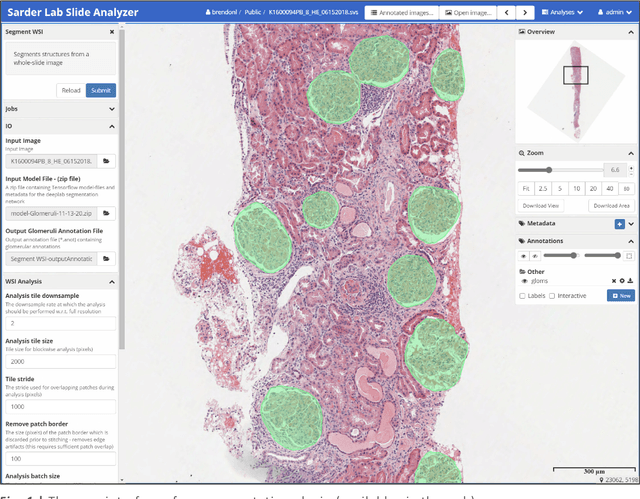
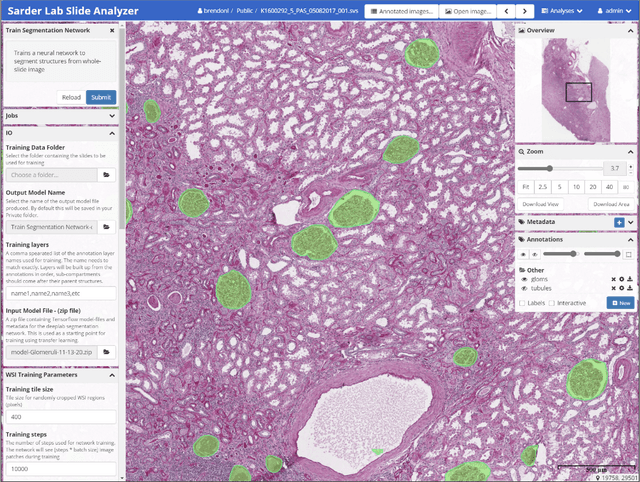
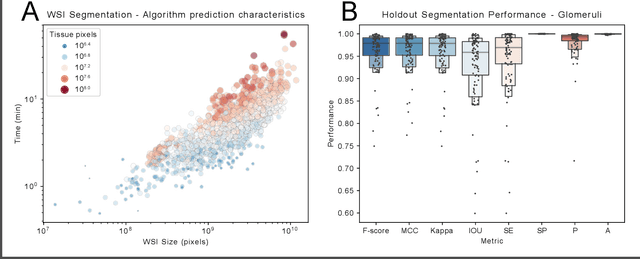
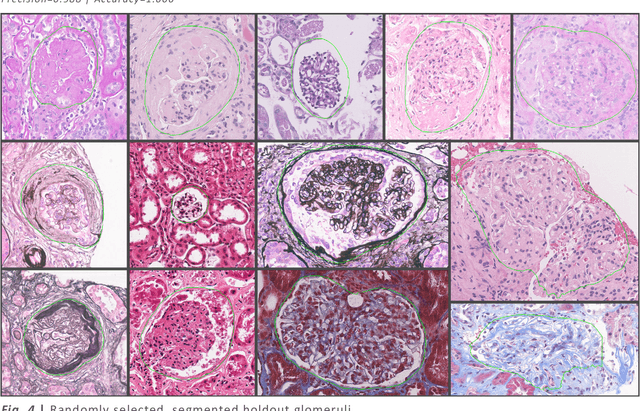
Abstract:Convolutional neural networks, the state of the art for image segmentation, have been successfully applied to histology images by many computational researchers. However, the translatability of this technology to clinicians and biological researchers is limited due to the complex and undeveloped user interface of the code, as well as the extensive computer setup required. As an extension of our previous work (arXiv:1812.07509), we have developed a tool for segmentation of whole slide images (WSIs) with an easy to use graphical user interface. Our tool runs a state-of-the-art convolutional neural network for segmentation of WSIs in the cloud. Our plugin is built on the open source tool HistomicsTK by Kitware Inc. (Clifton Park, NY), which provides remote data management and viewing abilities for WSI datasets. The ability to access this tool over the internet will facilitate widespread use by computational non-experts. Users can easily upload slides to a server where our plugin is installed and perform human in the loop segmentation analysis remotely. This tool is open source, and has the ability to be adapted to segment of any pathological structure. For a proof of concept, we have trained it to segment glomeruli from renal tissue images, achieving an F-score > 0.97 on holdout tissue slides.
Neural Network Segmentation of Interstitial Fibrosis, Tubular Atrophy, and Glomerulosclerosis in Renal Biopsies
Feb 28, 2020



Abstract:Glomerulosclerosis, interstitial fibrosis, and tubular atrophy (IFTA) are histologic indicators of irrecoverable kidney injury. In standard clinical practice, the renal pathologist visually assesses, under the microscope, the percentage of sclerotic glomeruli and the percentage of renal cortical involvement by IFTA. Estimation of IFTA is a subjective process due to a varied spectrum and definition of morphological manifestations. Modern artificial intelligence and computer vision algorithms have the ability to reduce inter-observer variability through rigorous quantitation. In this work, we apply convolutional neural networks for the segmentation of glomerulosclerosis and IFTA in periodic acid-Schiff stained renal biopsies. The convolutional network approach achieves high performance in intra-institutional holdout data, and achieves moderate performance in inter-intuitional holdout data, which the network had never seen in training. The convolutional approach demonstrated interesting properties, such as learning to predict regions better than the provided ground truth as well as developing its own conceptualization of segmental sclerosis. Subsequent estimations of IFTA and glomerulosclerosis percentages showed high correlation with ground truth.
Unsupervised Community Detection with a Potts Model Hamiltonian, an Efficient Algorithmic Solution, and Applications in Digital Pathology
Feb 05, 2020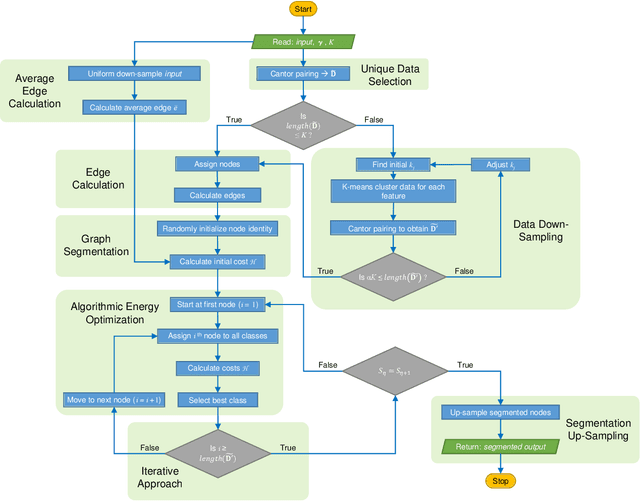
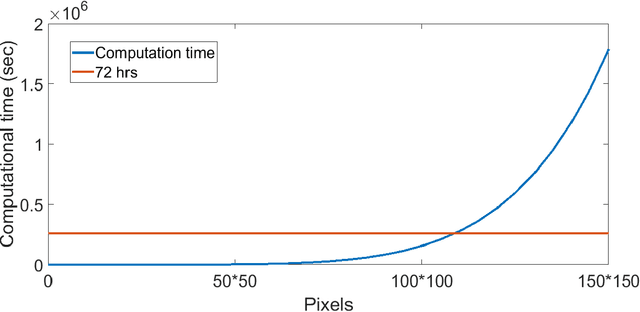
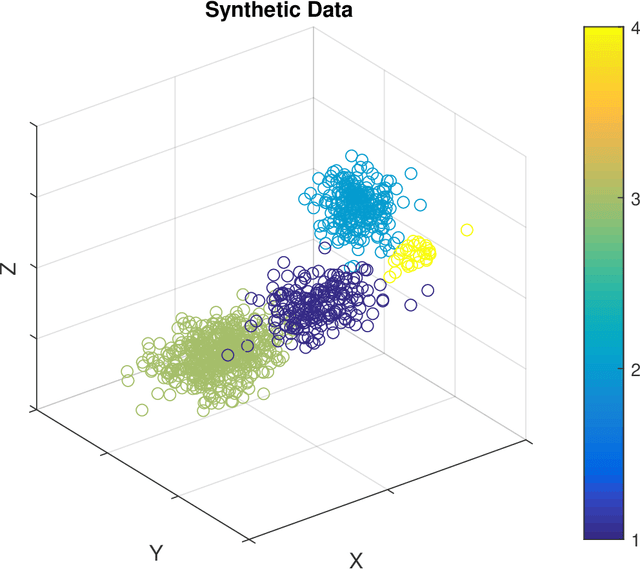
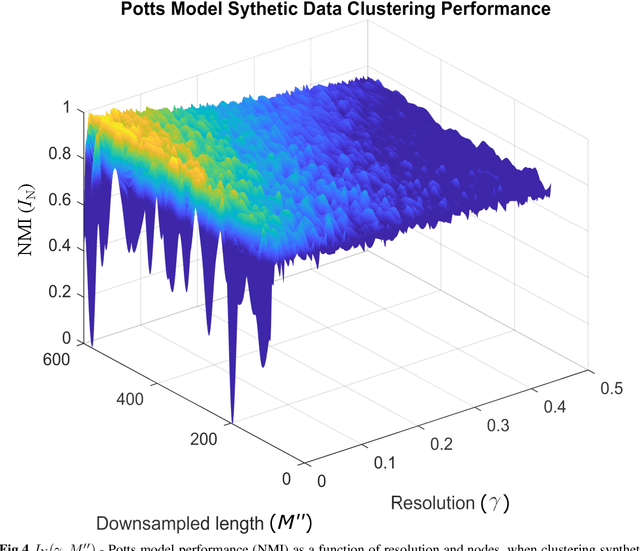
Abstract:Unsupervised segmentation of large images using a Potts model Hamiltonian is unique in that segmentation is governed by a resolution parameter which scales the sensitivity to small clusters. Here, the input image is first modeled as a graph, which is then segmented by minimizing a Hamiltonian cost function defined on the graph and the respective segments. However, there exists no closed form solution of this optimization, and using previous iterative algorithmic solution techniques, the problem scales quadratically in the Input Length. Therefore, while Potts model segmentation gives accurate segmentation, it is grossly underutilized as an unsupervised learning technique. We propose a fast statistical down-sampling of input image pixels based on the respective color features, and a new iterative method to minimize the Potts model energy considering pixel to segment relationship. This method is generalizable and can be extended for image pixel texture features as well as spatial features. We demonstrate that this new method is highly efficient, and outperforms existing methods for Potts model based image segmentation. We demonstrate the application of our method in medical microscopy image segmentation; particularly, in segmenting renal glomerular micro-environment in renal pathology. Our method is not limited to image segmentation, and can be extended to any image/data segmentation/clustering task for arbitrary datasets with discrete features.
Iterative annotation to ease neural network training: Specialized machine learning in medical image analysis
Dec 18, 2018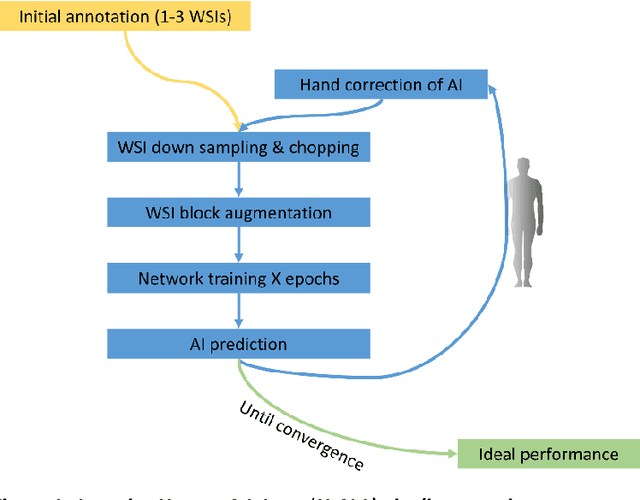
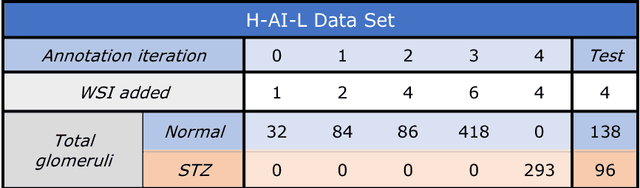
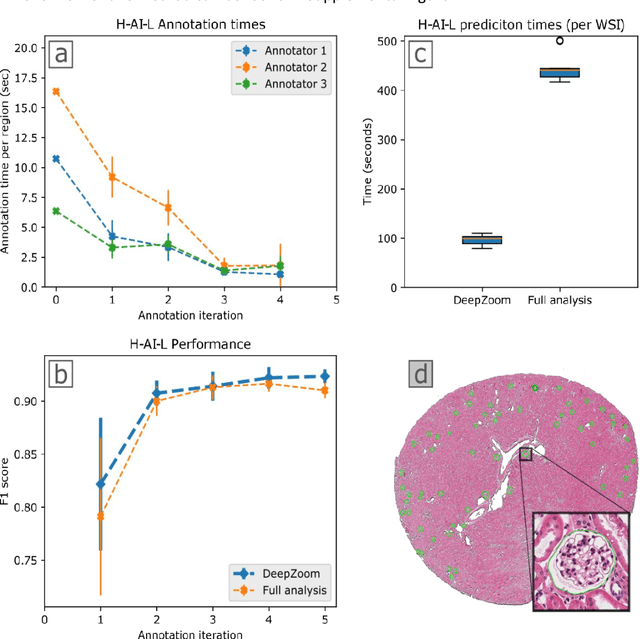
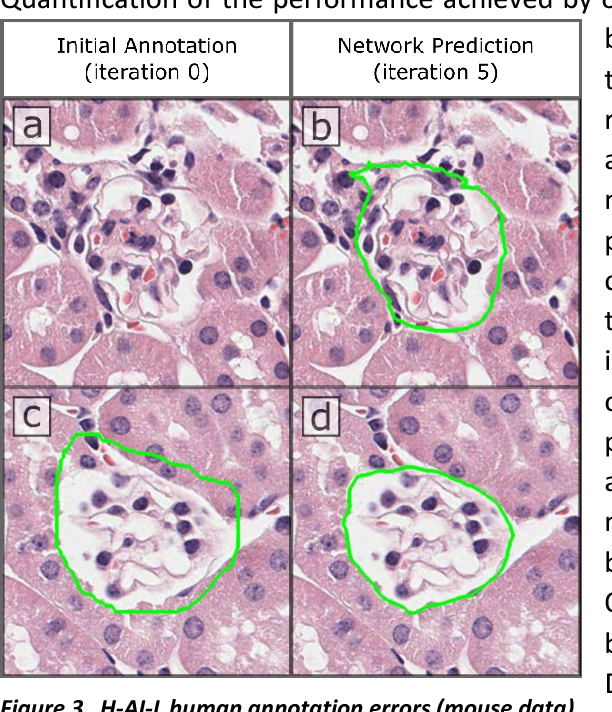
Abstract:Neural networks promise to bring robust, quantitative analysis to medical fields, but adoption is limited by the technicalities of training these networks. To address this translation gap between medical researchers and neural networks in the field of pathology, we have created an intuitive interface which utilizes the commonly used whole slide image (WSI) viewer, Aperio ImageScope (Leica Biosystems Imaging, Inc.), for the annotation and display of neural network predictions on WSIs. Leveraging this, we propose the use of a human-in-the-loop strategy to reduce the burden of WSI annotation. We track network performance improvements as a function of iteration and quantify the use of this pipeline for the segmentation of renal histologic findings on WSIs. More specifically, we present network performance when applied to segmentation of renal micro compartments, and demonstrate multi-class segmentation in human and mouse renal tissue slides. Finally, to show the adaptability of this technique to other medical imaging fields, we demonstrate its ability to iteratively segment human prostate glands from radiology imaging data.
Automatic Segmentation of Fluorescence Lifetime Microscopy Images of Cells Using Multi-Resolution Community Detection
May 08, 2013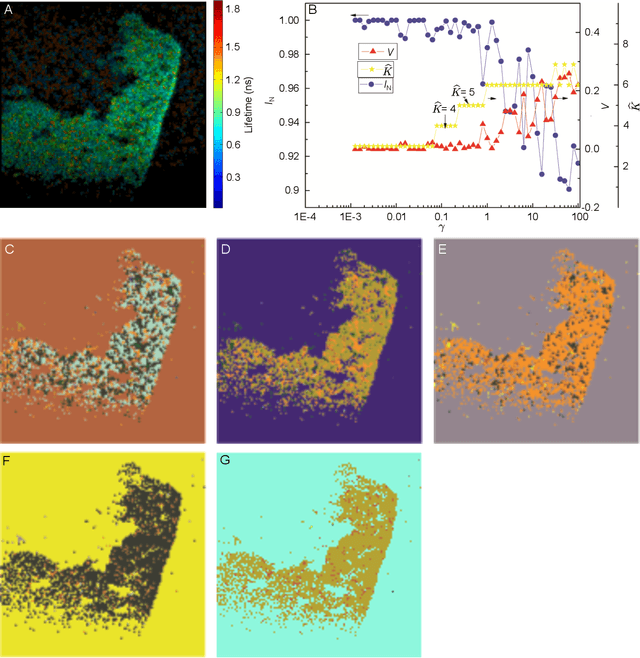


Abstract:We have developed an automatic method for segmenting fluorescence lifetime (FLT) imaging microscopy (FLIM) images of cells inspired by a multi-resolution community detection (MCD) based network segmentation method. The image processing problem is framed as identifying segments with respective average FLTs against a background in FLIM images. The proposed method segments a FLIM image for a given resolution of the network composed using image pixels as the nodes and similarity between the pixels as the edges. In the resulting segmentation, low network resolution leads to larger segments and high network resolution leads to smaller segments. Further, the mean-square error (MSE) in estimating the FLT segments in a FLIM image using the proposed method was found to be consistently decreasing with increasing resolution of the corresponding network. The proposed MCD method outperformed a popular spectral clustering based method in performing FLIM image segmentation. The spectral segmentation method introduced noisy segments in its output at high resolution. It was unable to offer a consistent decrease in MSE with increasing resolution.
* 21 pages, 6 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge