Kuang-Yu Jen
Department of Pathology and Laboratory Medicine, University of California, Davis Medical Center, Sacramento, California
Label-free evaluation of lung and heart transplant biopsies using virtual staining
Sep 09, 2024



Abstract:Organ transplantation serves as the primary therapeutic strategy for end-stage organ failures. However, allograft rejection is a common complication of organ transplantation. Histological assessment is essential for the timely detection and diagnosis of transplant rejection and remains the gold standard. Nevertheless, the traditional histochemical staining process is time-consuming, costly, and labor-intensive. Here, we present a panel of virtual staining neural networks for lung and heart transplant biopsies, which digitally convert autofluorescence microscopic images of label-free tissue sections into their brightfield histologically stained counterparts, bypassing the traditional histochemical staining process. Specifically, we virtually generated Hematoxylin and Eosin (H&E), Masson's Trichrome (MT), and Elastic Verhoeff-Van Gieson (EVG) stains for label-free transplant lung tissue, along with H&E and MT stains for label-free transplant heart tissue. Subsequent blind evaluations conducted by three board-certified pathologists have confirmed that the virtual staining networks consistently produce high-quality histology images with high color uniformity, closely resembling their well-stained histochemical counterparts across various tissue features. The use of virtually stained images for the evaluation of transplant biopsies achieved comparable diagnostic outcomes to those obtained via traditional histochemical staining, with a concordance rate of 82.4% for lung samples and 91.7% for heart samples. Moreover, virtual staining models create multiple stains from the same autofluorescence input, eliminating structural mismatches observed between adjacent sections stained in the traditional workflow, while also saving tissue, expert time, and staining costs.
Neural Network Segmentation of Interstitial Fibrosis, Tubular Atrophy, and Glomerulosclerosis in Renal Biopsies
Feb 28, 2020



Abstract:Glomerulosclerosis, interstitial fibrosis, and tubular atrophy (IFTA) are histologic indicators of irrecoverable kidney injury. In standard clinical practice, the renal pathologist visually assesses, under the microscope, the percentage of sclerotic glomeruli and the percentage of renal cortical involvement by IFTA. Estimation of IFTA is a subjective process due to a varied spectrum and definition of morphological manifestations. Modern artificial intelligence and computer vision algorithms have the ability to reduce inter-observer variability through rigorous quantitation. In this work, we apply convolutional neural networks for the segmentation of glomerulosclerosis and IFTA in periodic acid-Schiff stained renal biopsies. The convolutional network approach achieves high performance in intra-institutional holdout data, and achieves moderate performance in inter-intuitional holdout data, which the network had never seen in training. The convolutional approach demonstrated interesting properties, such as learning to predict regions better than the provided ground truth as well as developing its own conceptualization of segmental sclerosis. Subsequent estimations of IFTA and glomerulosclerosis percentages showed high correlation with ground truth.
Iterative annotation to ease neural network training: Specialized machine learning in medical image analysis
Dec 18, 2018
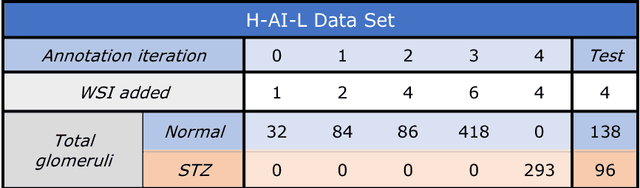
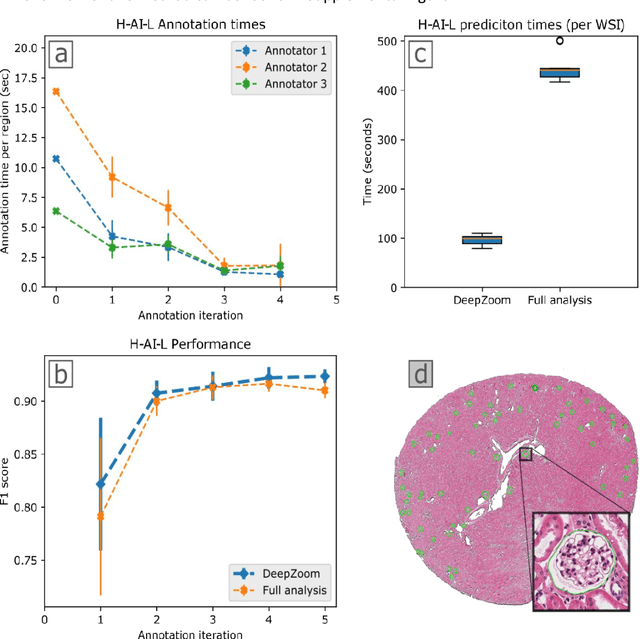
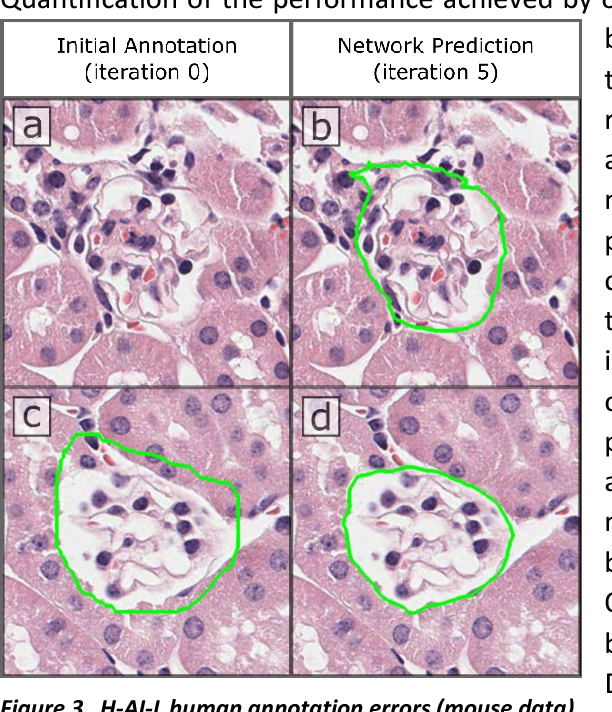
Abstract:Neural networks promise to bring robust, quantitative analysis to medical fields, but adoption is limited by the technicalities of training these networks. To address this translation gap between medical researchers and neural networks in the field of pathology, we have created an intuitive interface which utilizes the commonly used whole slide image (WSI) viewer, Aperio ImageScope (Leica Biosystems Imaging, Inc.), for the annotation and display of neural network predictions on WSIs. Leveraging this, we propose the use of a human-in-the-loop strategy to reduce the burden of WSI annotation. We track network performance improvements as a function of iteration and quantify the use of this pipeline for the segmentation of renal histologic findings on WSIs. More specifically, we present network performance when applied to segmentation of renal micro compartments, and demonstrate multi-class segmentation in human and mouse renal tissue slides. Finally, to show the adaptability of this technique to other medical imaging fields, we demonstrate its ability to iteratively segment human prostate glands from radiology imaging data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge