Brendon Lutnick
PSO-Net: Development of an automated psoriasis assessment system using attention-based interpretable deep neural networks
Jan 30, 2025Abstract:Psoriasis is a chronic skin condition that requires long-term treatment and monitoring. Although, the Psoriasis Area and Severity Index (PASI) is utilized as a standard measurement to assess psoriasis severity in clinical trials, it has many drawbacks such as (1) patient burden for in-person clinic visits for assessment of psoriasis, (2) time required for investigator scoring and (3) variability of inter- and intra-rater scoring. To address these drawbacks, we propose a novel and interpretable deep learning architecture called PSO-Net, which maps digital images from different anatomical regions to derive attention-based scores. Regional scores are further combined to estimate an absolute PASI score. Moreover, we devise a novel regression activation map for interpretability through ranking attention scores. Using this approach, we achieved inter-class correlation scores of 82.2% [95% CI: 77- 87%] and 87.8% [95% CI: 84-91%] with two different clinician raters, respectively.
Histo-fetch -- On-the-fly processing of gigapixel whole slide images simplifies and speeds neural network training
Mar 01, 2021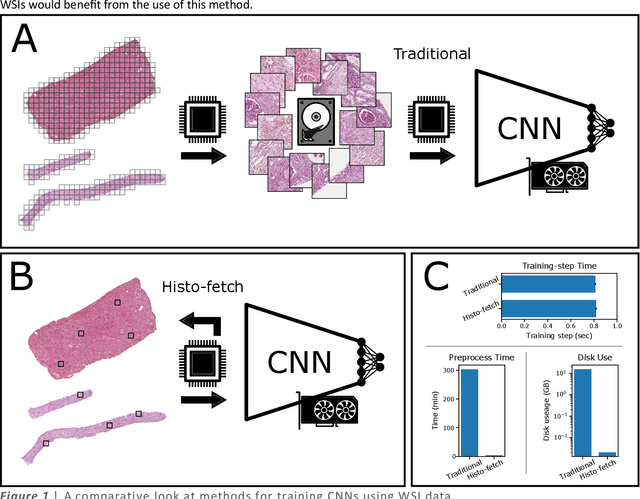

Abstract:We created a custom pipeline (histo-fetch) to efficiently extract random patches and labels from pathology whole slide images (WSIs) for input to a neural network on-the-fly. We prefetch these patches as needed during network training, avoiding the need for WSI preparation such as chopping/tiling. We demonstrate the utility of this pipeline to perform artificial stain transfer and image generation using the popular networks CycleGAN and ProGAN, respectively.
A tool for user friendly, cloud based, whole slide image segmentation
Jan 18, 2021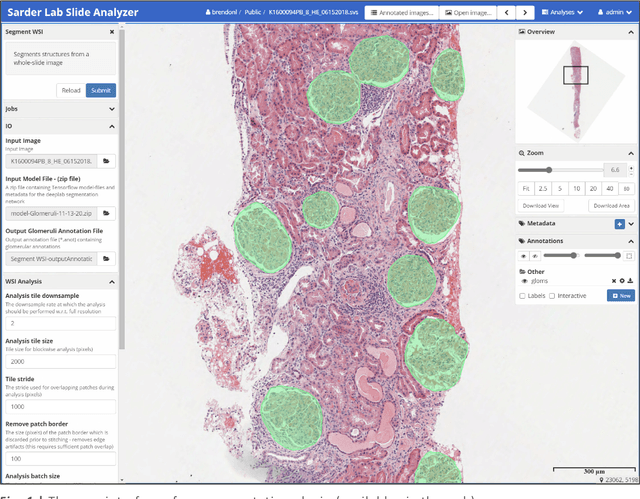
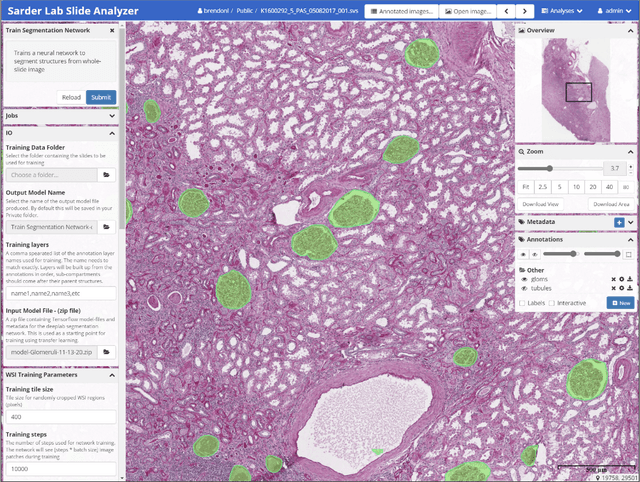
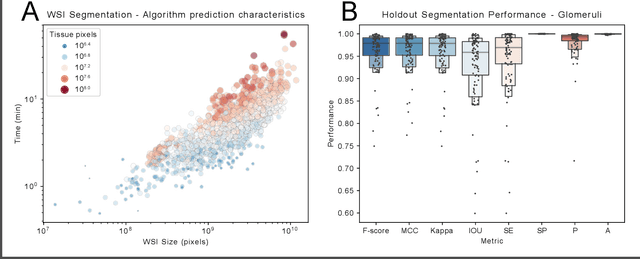
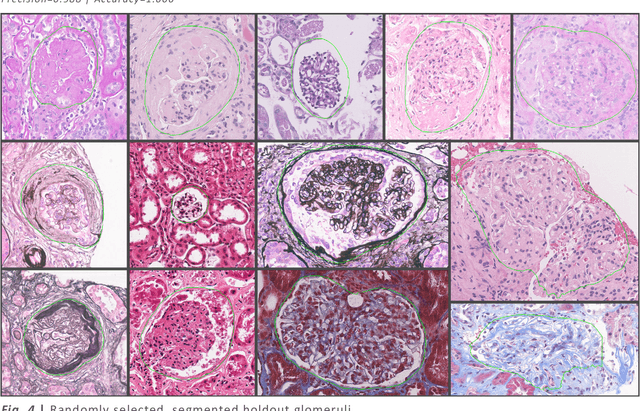
Abstract:Convolutional neural networks, the state of the art for image segmentation, have been successfully applied to histology images by many computational researchers. However, the translatability of this technology to clinicians and biological researchers is limited due to the complex and undeveloped user interface of the code, as well as the extensive computer setup required. As an extension of our previous work (arXiv:1812.07509), we have developed a tool for segmentation of whole slide images (WSIs) with an easy to use graphical user interface. Our tool runs a state-of-the-art convolutional neural network for segmentation of WSIs in the cloud. Our plugin is built on the open source tool HistomicsTK by Kitware Inc. (Clifton Park, NY), which provides remote data management and viewing abilities for WSI datasets. The ability to access this tool over the internet will facilitate widespread use by computational non-experts. Users can easily upload slides to a server where our plugin is installed and perform human in the loop segmentation analysis remotely. This tool is open source, and has the ability to be adapted to segment of any pathological structure. For a proof of concept, we have trained it to segment glomeruli from renal tissue images, achieving an F-score > 0.97 on holdout tissue slides.
Unsupervised Community Detection with a Potts Model Hamiltonian, an Efficient Algorithmic Solution, and Applications in Digital Pathology
Feb 05, 2020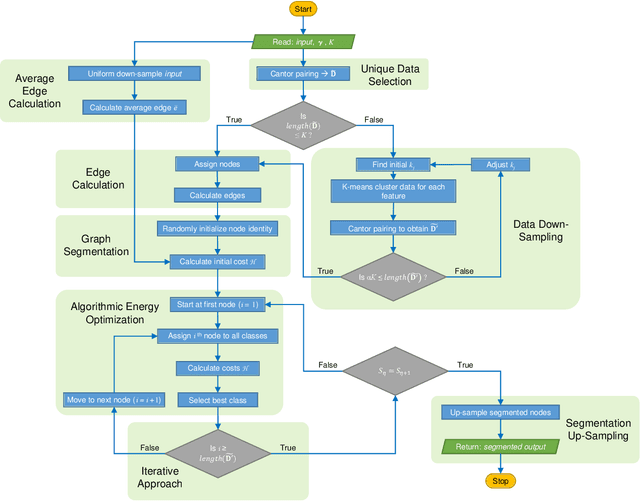
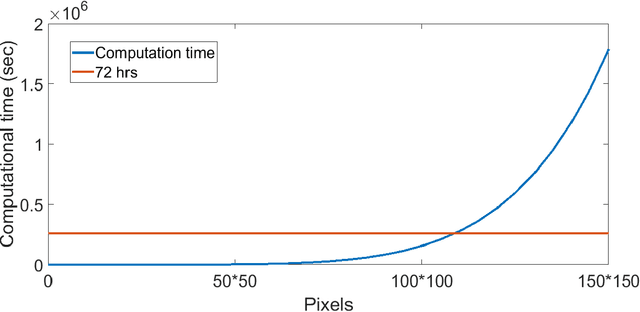
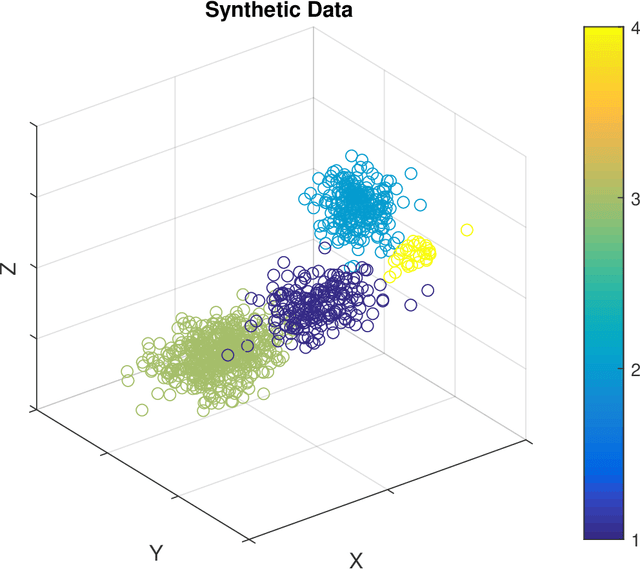
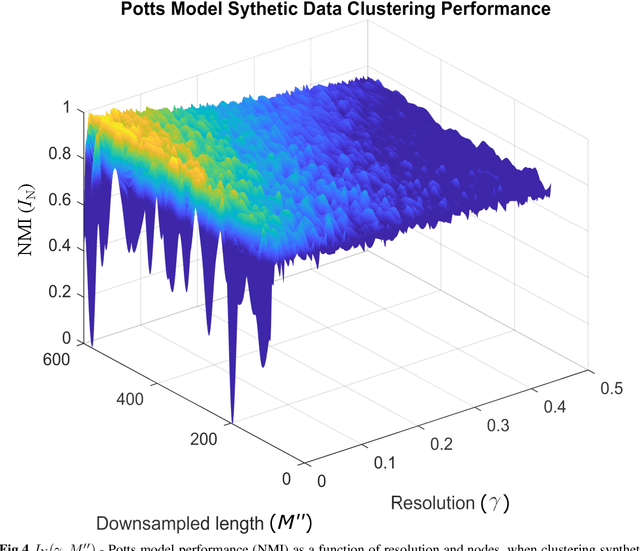
Abstract:Unsupervised segmentation of large images using a Potts model Hamiltonian is unique in that segmentation is governed by a resolution parameter which scales the sensitivity to small clusters. Here, the input image is first modeled as a graph, which is then segmented by minimizing a Hamiltonian cost function defined on the graph and the respective segments. However, there exists no closed form solution of this optimization, and using previous iterative algorithmic solution techniques, the problem scales quadratically in the Input Length. Therefore, while Potts model segmentation gives accurate segmentation, it is grossly underutilized as an unsupervised learning technique. We propose a fast statistical down-sampling of input image pixels based on the respective color features, and a new iterative method to minimize the Potts model energy considering pixel to segment relationship. This method is generalizable and can be extended for image pixel texture features as well as spatial features. We demonstrate that this new method is highly efficient, and outperforms existing methods for Potts model based image segmentation. We demonstrate the application of our method in medical microscopy image segmentation; particularly, in segmenting renal glomerular micro-environment in renal pathology. Our method is not limited to image segmentation, and can be extended to any image/data segmentation/clustering task for arbitrary datasets with discrete features.
Iterative annotation to ease neural network training: Specialized machine learning in medical image analysis
Dec 18, 2018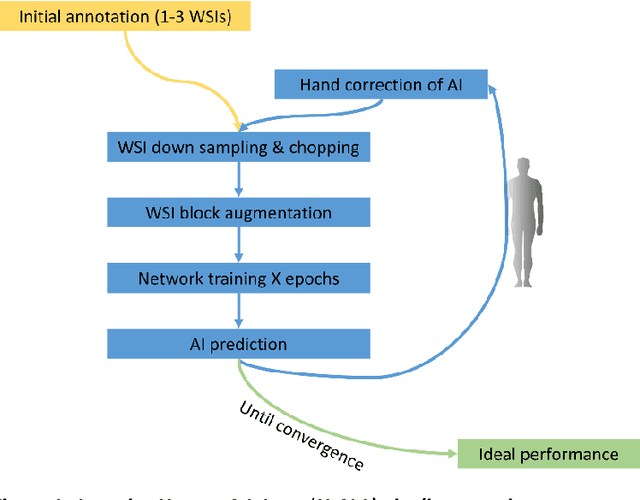
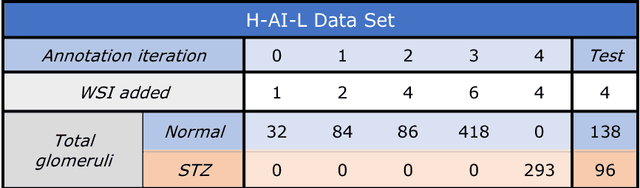
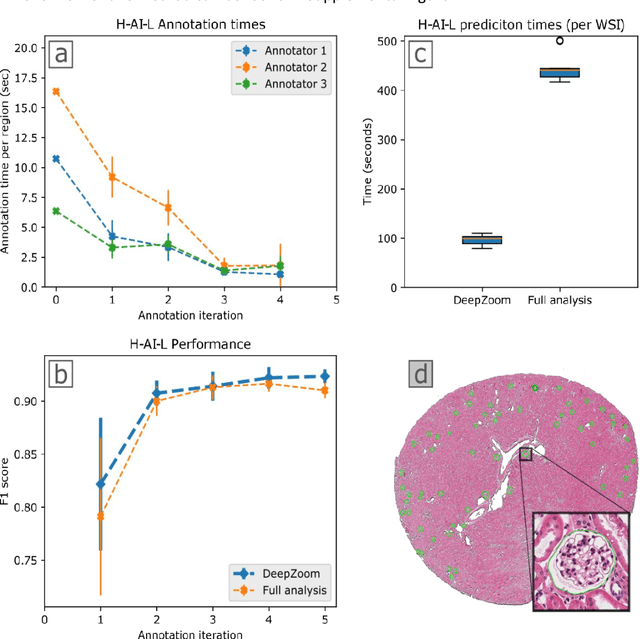
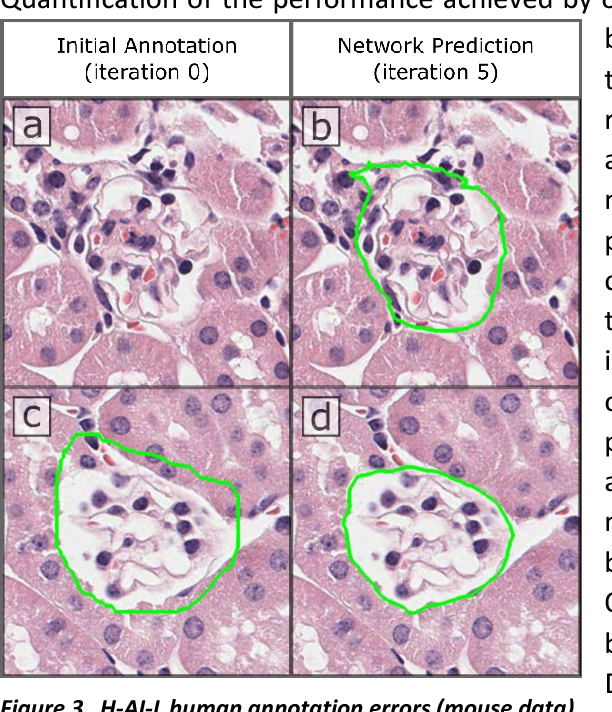
Abstract:Neural networks promise to bring robust, quantitative analysis to medical fields, but adoption is limited by the technicalities of training these networks. To address this translation gap between medical researchers and neural networks in the field of pathology, we have created an intuitive interface which utilizes the commonly used whole slide image (WSI) viewer, Aperio ImageScope (Leica Biosystems Imaging, Inc.), for the annotation and display of neural network predictions on WSIs. Leveraging this, we propose the use of a human-in-the-loop strategy to reduce the burden of WSI annotation. We track network performance improvements as a function of iteration and quantify the use of this pipeline for the segmentation of renal histologic findings on WSIs. More specifically, we present network performance when applied to segmentation of renal micro compartments, and demonstrate multi-class segmentation in human and mouse renal tissue slides. Finally, to show the adaptability of this technique to other medical imaging fields, we demonstrate its ability to iteratively segment human prostate glands from radiology imaging data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge