Brandon Ginley
Department of Pathology & Anatomical Sciences, University at Buffalo, the State University of New York, Buffalo, New York
Histo-fetch -- On-the-fly processing of gigapixel whole slide images simplifies and speeds neural network training
Mar 01, 2021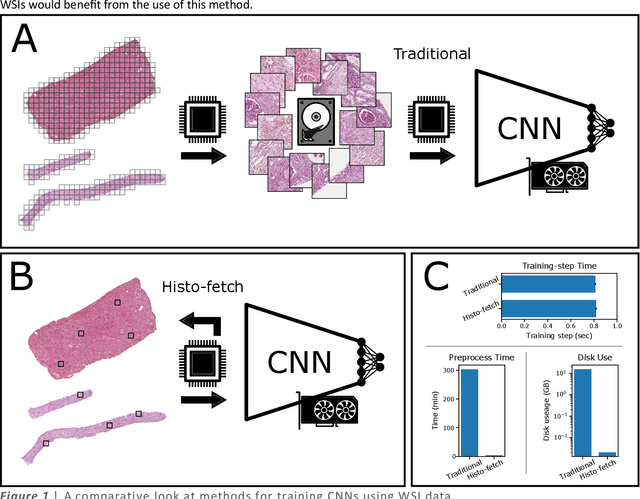

Abstract:We created a custom pipeline (histo-fetch) to efficiently extract random patches and labels from pathology whole slide images (WSIs) for input to a neural network on-the-fly. We prefetch these patches as needed during network training, avoiding the need for WSI preparation such as chopping/tiling. We demonstrate the utility of this pipeline to perform artificial stain transfer and image generation using the popular networks CycleGAN and ProGAN, respectively.
Neural Network Segmentation of Interstitial Fibrosis, Tubular Atrophy, and Glomerulosclerosis in Renal Biopsies
Feb 28, 2020



Abstract:Glomerulosclerosis, interstitial fibrosis, and tubular atrophy (IFTA) are histologic indicators of irrecoverable kidney injury. In standard clinical practice, the renal pathologist visually assesses, under the microscope, the percentage of sclerotic glomeruli and the percentage of renal cortical involvement by IFTA. Estimation of IFTA is a subjective process due to a varied spectrum and definition of morphological manifestations. Modern artificial intelligence and computer vision algorithms have the ability to reduce inter-observer variability through rigorous quantitation. In this work, we apply convolutional neural networks for the segmentation of glomerulosclerosis and IFTA in periodic acid-Schiff stained renal biopsies. The convolutional network approach achieves high performance in intra-institutional holdout data, and achieves moderate performance in inter-intuitional holdout data, which the network had never seen in training. The convolutional approach demonstrated interesting properties, such as learning to predict regions better than the provided ground truth as well as developing its own conceptualization of segmental sclerosis. Subsequent estimations of IFTA and glomerulosclerosis percentages showed high correlation with ground truth.
Iterative annotation to ease neural network training: Specialized machine learning in medical image analysis
Dec 18, 2018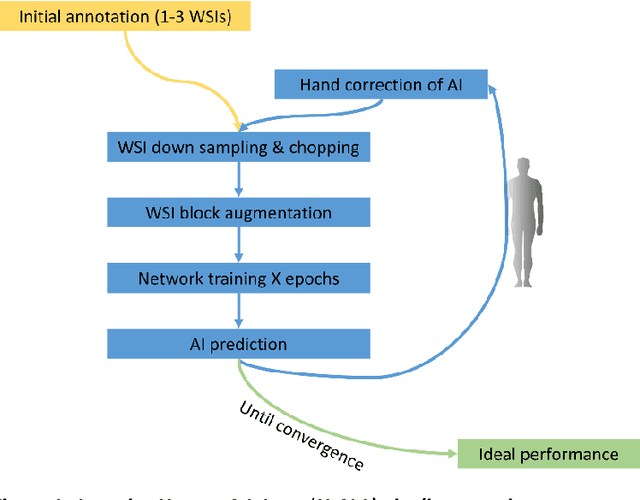
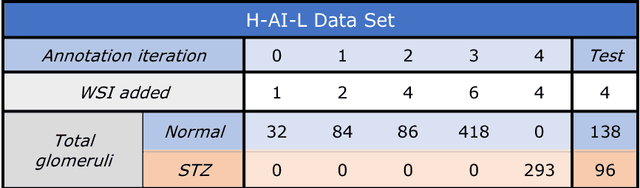
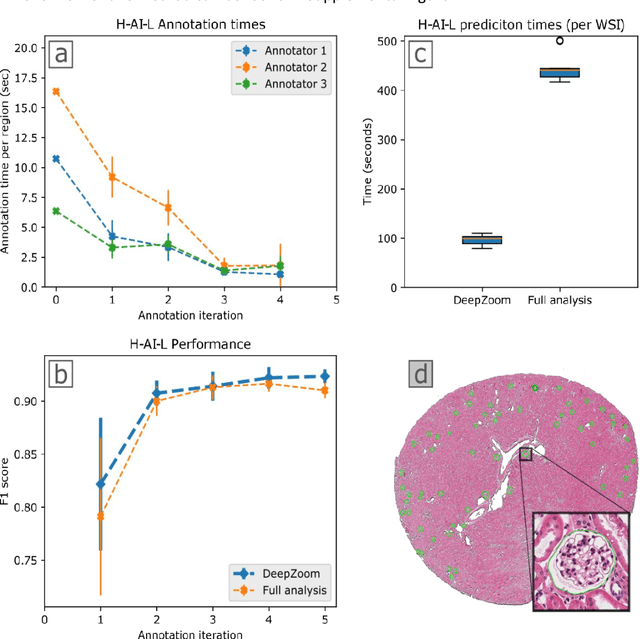
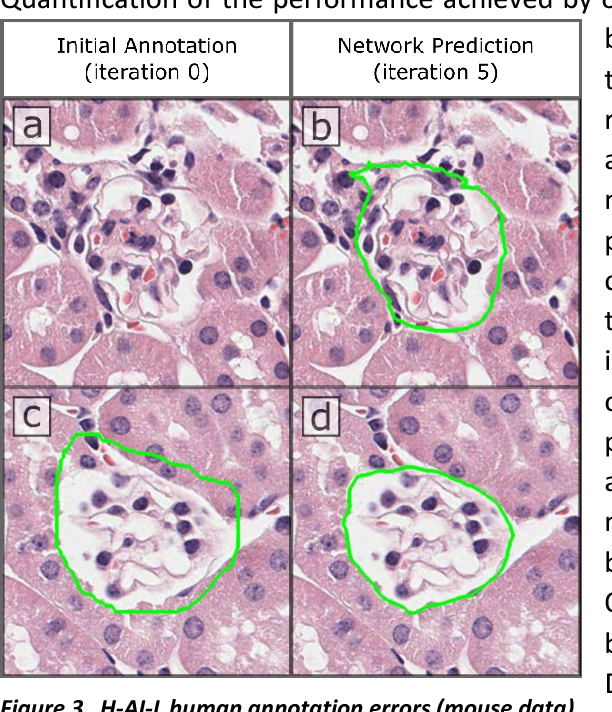
Abstract:Neural networks promise to bring robust, quantitative analysis to medical fields, but adoption is limited by the technicalities of training these networks. To address this translation gap between medical researchers and neural networks in the field of pathology, we have created an intuitive interface which utilizes the commonly used whole slide image (WSI) viewer, Aperio ImageScope (Leica Biosystems Imaging, Inc.), for the annotation and display of neural network predictions on WSIs. Leveraging this, we propose the use of a human-in-the-loop strategy to reduce the burden of WSI annotation. We track network performance improvements as a function of iteration and quantify the use of this pipeline for the segmentation of renal histologic findings on WSIs. More specifically, we present network performance when applied to segmentation of renal micro compartments, and demonstrate multi-class segmentation in human and mouse renal tissue slides. Finally, to show the adaptability of this technique to other medical imaging fields, we demonstrate its ability to iteratively segment human prostate glands from radiology imaging data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge