Philippe A. Robert
Opponent Shaping for Antibody Development
Sep 19, 2024Abstract:Anti-viral therapies are typically designed or evolved towards the current strains of a virus. In learning terms, this corresponds to a myopic best response, i.e., not considering the possible adaptive moves of the opponent. However, therapy-induced selective pressures act on viral antigens to drive the emergence of mutated strains, against which initial therapies have reduced efficacy. To motivate our work, we consider antibody designs that target not only the current viral strains but also the wide range of possible future variants that the virus might evolve into under the evolutionary pressure exerted by said antibodies. Building on a computational model of binding between antibodies and viral antigens (the Absolut! framework), we design and implement a genetic simulation of the viral evolutionary escape. Crucially, this allows our antibody optimisation algorithm to consider and influence the entire escape curve of the virus, i.e. to guide (or ''shape'') the viral evolution. This is inspired by opponent shaping which, in general-sum learning, accounts for the adaptation of the co-player rather than playing a myopic best response. Hence we call the optimised antibodies shapers. Within our simulations, we demonstrate that our shapers target both current and simulated future viral variants, outperforming the antibodies chosen in a myopic way. Furthermore, we show that shapers exert specific evolutionary pressure on the virus compared to myopic antibodies. Altogether, shapers modify the evolutionary trajectories of viral strains and minimise the viral escape compared to their myopic counterparts. While this is a simple model, we hope that our proposed paradigm will enable the discovery of better long-lived vaccines and antibody therapies in the future, enabled by rapid advancements in the capabilities of simulation tools.
ImmunoLingo: Linguistics-based formalization of the antibody language
Sep 26, 2022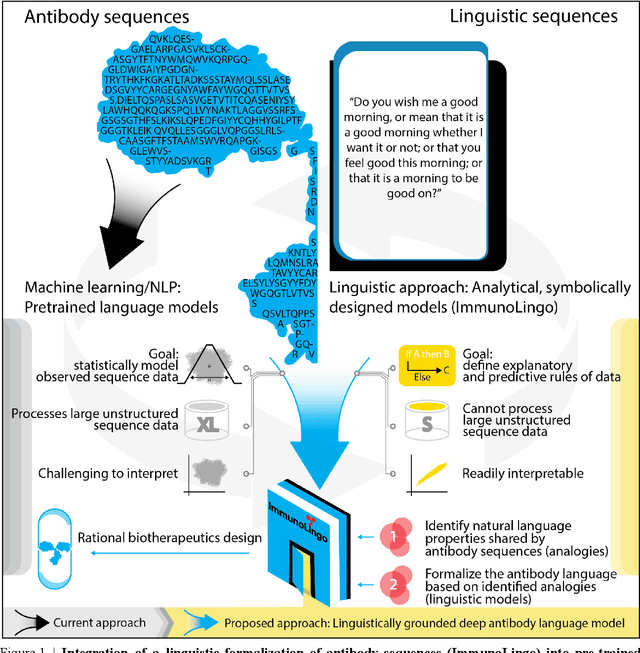
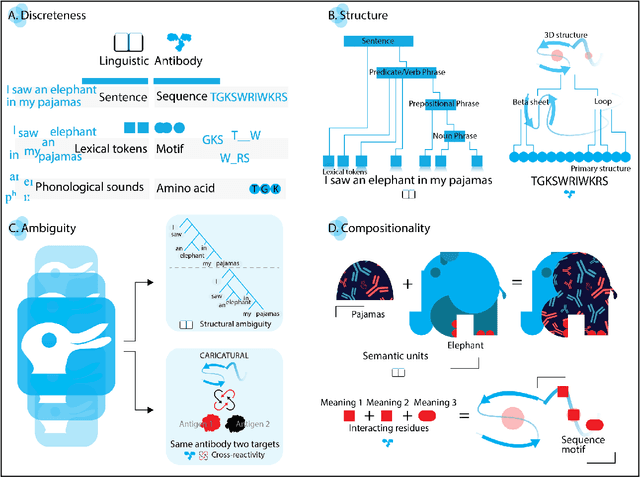
Abstract:Apparent parallels between natural language and biological sequence have led to a recent surge in the application of deep language models (LMs) to the analysis of antibody and other biological sequences. However, a lack of a rigorous linguistic formalization of biological sequence languages, which would define basic components, such as lexicon (i.e., the discrete units of the language) and grammar (i.e., the rules that link sequence well-formedness, structure, and meaning) has led to largely domain-unspecific applications of LMs, which do not take into account the underlying structure of the biological sequences studied. A linguistic formalization, on the other hand, establishes linguistically-informed and thus domain-adapted components for LM applications. It would facilitate a better understanding of how differences and similarities between natural language and biological sequences influence the quality of LMs, which is crucial for the design of interpretable models with extractable sequence-functions relationship rules, such as the ones underlying the antibody specificity prediction problem. Deciphering the rules of antibody specificity is crucial to accelerating rational and in silico biotherapeutic drug design. Here, we formalize the properties of the antibody language and thereby establish not only a foundation for the application of linguistic tools in adaptive immune receptor analysis but also for the systematic immunolinguistic studies of immune receptor specificity in general.
Advancing protein language models with linguistics: a roadmap for improved interpretability
Jul 03, 2022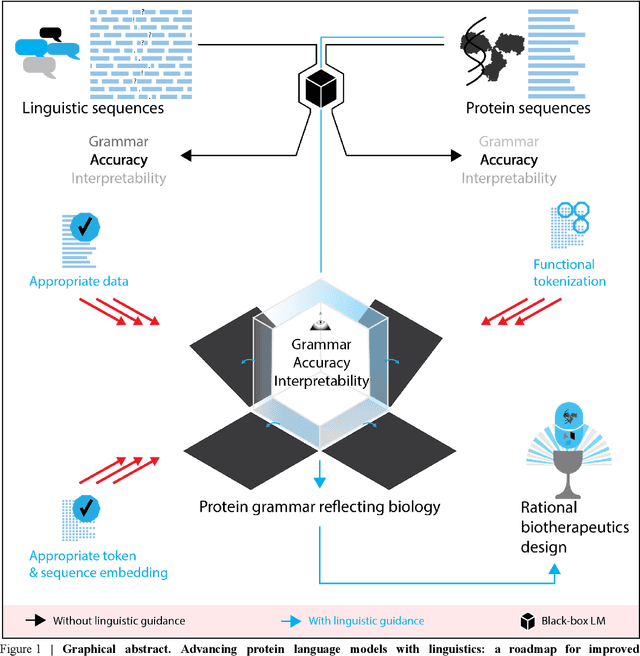
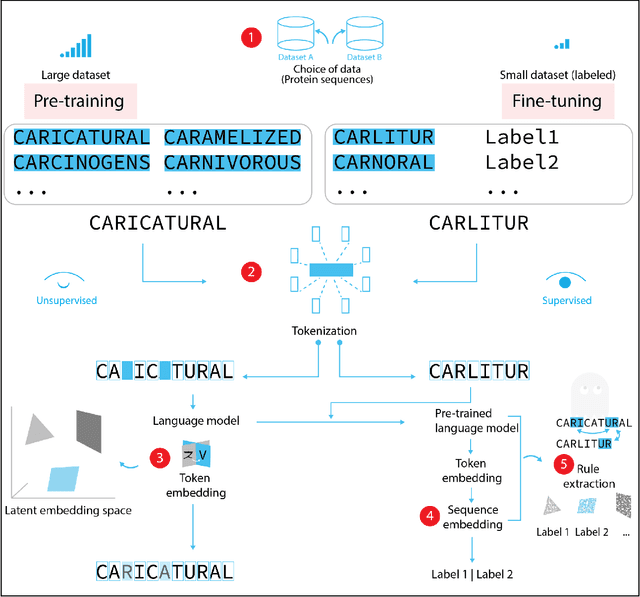
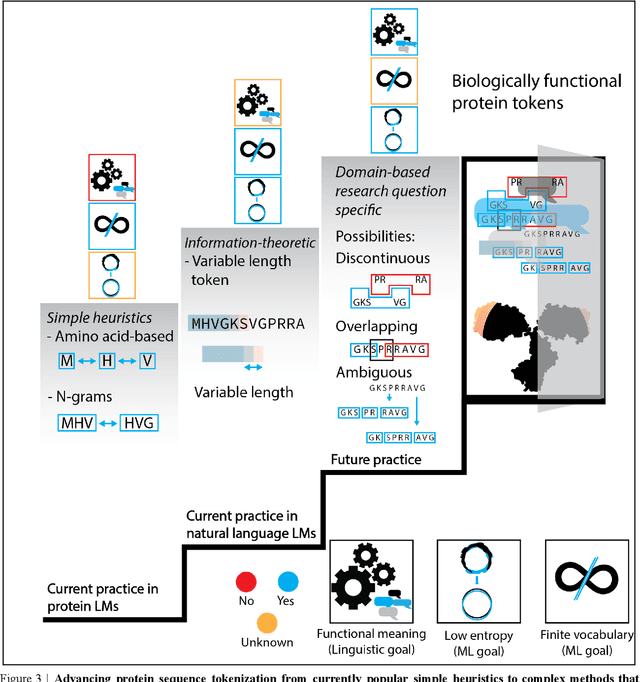
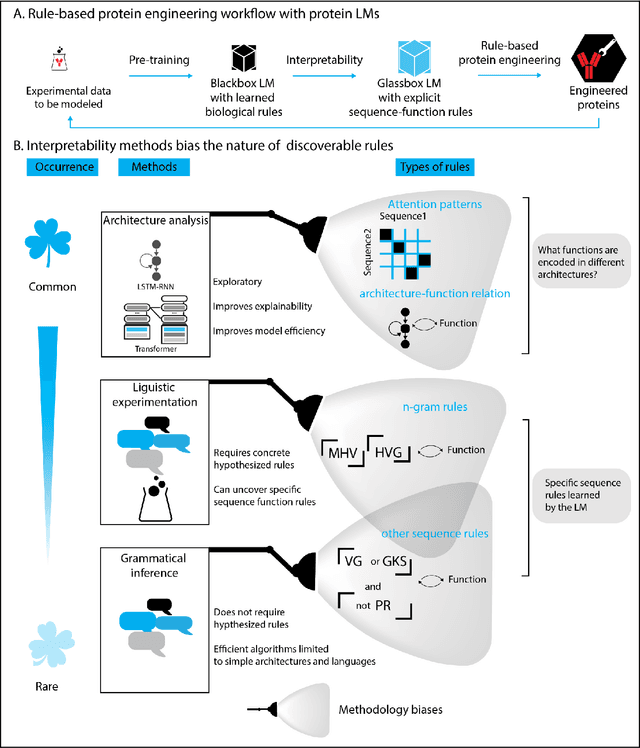
Abstract:Deep neural-network-based language models (LMs) are increasingly applied to large-scale protein sequence data to predict protein function. However, being largely blackbox models and thus challenging to interpret, current protein LM approaches do not contribute to a fundamental understanding of sequence-function mappings, hindering rule-based biotherapeutic drug development. We argue that guidance drawn from linguistics, a field specialized in analytical rule extraction from natural language data, can aid with building more interpretable protein LMs that have learned relevant domain-specific rules. Differences between protein sequence data and linguistic sequence data require the integration of more domain-specific knowledge in protein LMs compared to natural language LMs. Here, we provide a linguistics-based roadmap for protein LM pipeline choices with regard to training data, tokenization, token embedding, sequence embedding, and model interpretation. Combining linguistics with protein LMs enables the development of next-generation interpretable machine learning models with the potential of uncovering the biological mechanisms underlying sequence-function relationships.
AntBO: Towards Real-World Automated Antibody Design with Combinatorial Bayesian Optimisation
Feb 16, 2022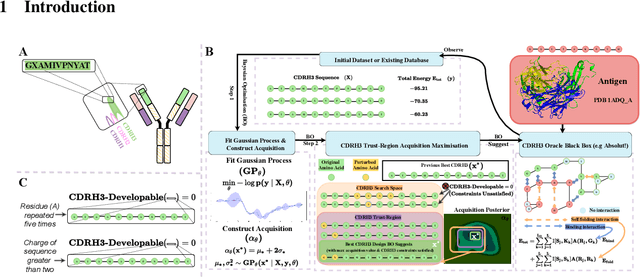
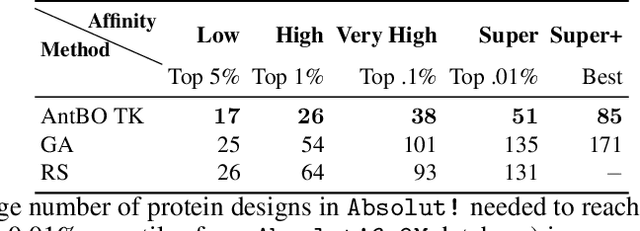
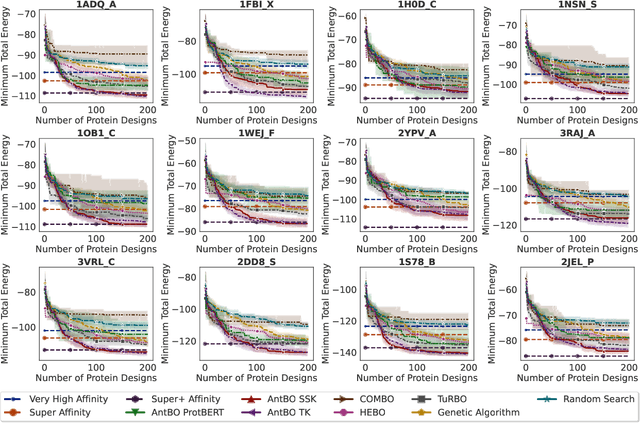
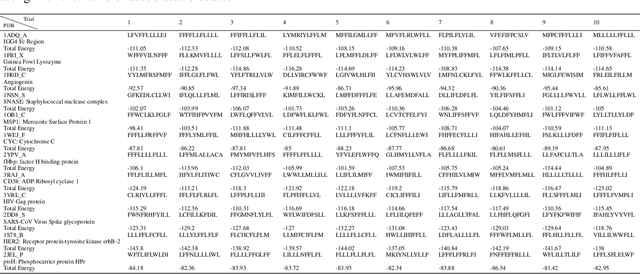
Abstract:Antibodies are canonically Y-shaped multimeric proteins capable of highly specific molecular recognition. The CDRH3 region located at the tip of variable chains of an antibody dominates antigen-binding specificity. Therefore, it is a priority to design optimal antigen-specific CDRH3 regions to develop therapeutic antibodies to combat harmful pathogens. However, the combinatorial nature of CDRH3 sequence space makes it impossible to search for an optimal binding sequence exhaustively and efficiently, especially not experimentally. Here, we present AntBO: a Combinatorial Bayesian Optimisation framework enabling efficient in silico design of the CDRH3 region. Ideally, antibodies should bind to their target antigen and be free from any harmful outcomes. Therefore, we introduce the CDRH3 trust region that restricts the search to sequences with feasible developability scores. To benchmark AntBO, we use the Absolut! software suite as a black-box oracle because it can score the target specificity and affinity of designed antibodies in silico in an unconstrained fashion. The results across 188 antigens demonstrate the benefit of AntBO in designing CDRH3 regions with diverse biophysical properties. In under 200 protein designs, AntBO can suggest antibody sequences that outperform the best binding sequence drawn from 6.9 million experimentally obtained CDRH3s and a commonly used genetic algorithm baseline. Additionally, AntBO finds very-high affinity CDRH3 sequences in only 38 protein designs whilst requiring no domain knowledge. We conclude AntBO brings automated antibody design methods closer to what is practically viable for in vitro experimentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge