Dany Bou-Ammar
AntBO: Towards Real-World Automated Antibody Design with Combinatorial Bayesian Optimisation
Feb 16, 2022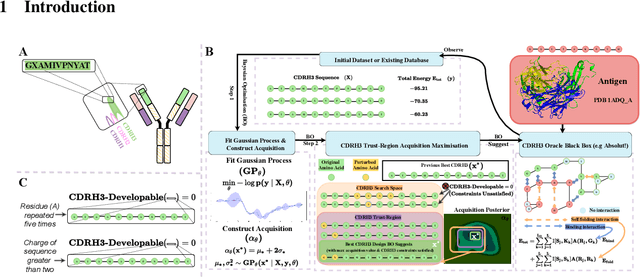
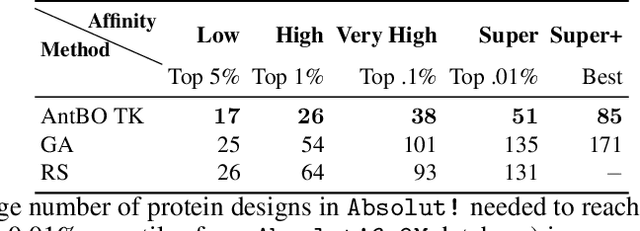
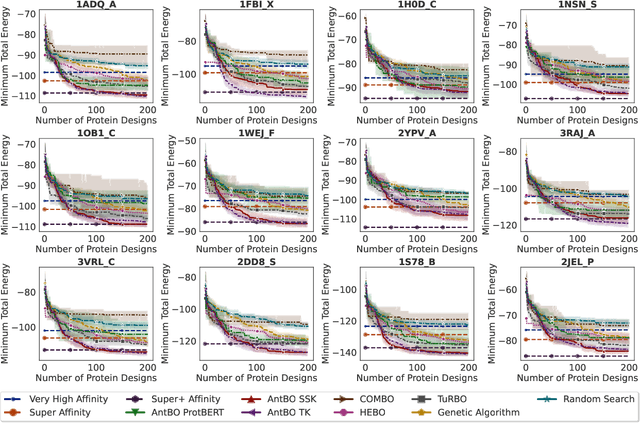
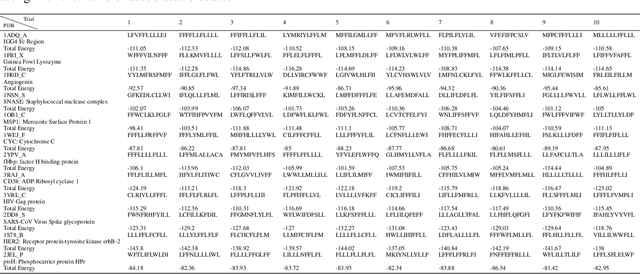
Abstract:Antibodies are canonically Y-shaped multimeric proteins capable of highly specific molecular recognition. The CDRH3 region located at the tip of variable chains of an antibody dominates antigen-binding specificity. Therefore, it is a priority to design optimal antigen-specific CDRH3 regions to develop therapeutic antibodies to combat harmful pathogens. However, the combinatorial nature of CDRH3 sequence space makes it impossible to search for an optimal binding sequence exhaustively and efficiently, especially not experimentally. Here, we present AntBO: a Combinatorial Bayesian Optimisation framework enabling efficient in silico design of the CDRH3 region. Ideally, antibodies should bind to their target antigen and be free from any harmful outcomes. Therefore, we introduce the CDRH3 trust region that restricts the search to sequences with feasible developability scores. To benchmark AntBO, we use the Absolut! software suite as a black-box oracle because it can score the target specificity and affinity of designed antibodies in silico in an unconstrained fashion. The results across 188 antigens demonstrate the benefit of AntBO in designing CDRH3 regions with diverse biophysical properties. In under 200 protein designs, AntBO can suggest antibody sequences that outperform the best binding sequence drawn from 6.9 million experimentally obtained CDRH3s and a commonly used genetic algorithm baseline. Additionally, AntBO finds very-high affinity CDRH3 sequences in only 38 protein designs whilst requiring no domain knowledge. We conclude AntBO brings automated antibody design methods closer to what is practically viable for in vitro experimentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge