Peng Qiu
ERNIE 5.0 Technical Report
Feb 04, 2026Abstract:In this report, we introduce ERNIE 5.0, a natively autoregressive foundation model desinged for unified multimodal understanding and generation across text, image, video, and audio. All modalities are trained from scratch under a unified next-group-of-tokens prediction objective, based on an ultra-sparse mixture-of-experts (MoE) architecture with modality-agnostic expert routing. To address practical challenges in large-scale deployment under diverse resource constraints, ERNIE 5.0 adopts a novel elastic training paradigm. Within a single pre-training run, the model learns a family of sub-models with varying depths, expert capacities, and routing sparsity, enabling flexible trade-offs among performance, model size, and inference latency in memory- or time-constrained scenarios. Moreover, we systematically address the challenges of scaling reinforcement learning to unified foundation models, thereby guaranteeing efficient and stable post-training under ultra-sparse MoE architectures and diverse multimodal settings. Extensive experiments demonstrate that ERNIE 5.0 achieves strong and balanced performance across multiple modalities. To the best of our knowledge, among publicly disclosed models, ERNIE 5.0 represents the first production-scale realization of a trillion-parameter unified autoregressive model that supports both multimodal understanding and generation. To facilitate further research, we present detailed visualizations of modality-agnostic expert routing in the unified model, alongside comprehensive empirical analysis of elastic training, aiming to offer profound insights to the community.
MobRT: A Digital Twin-Based Framework for Scalable Learning in Mobile Manipulation
Oct 06, 2025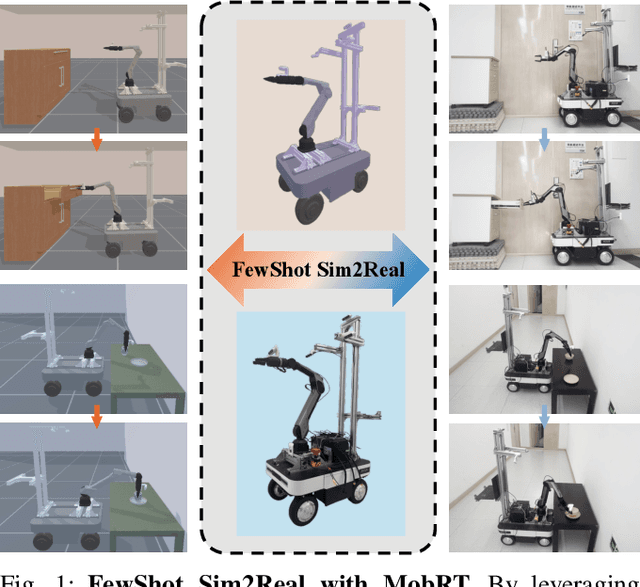
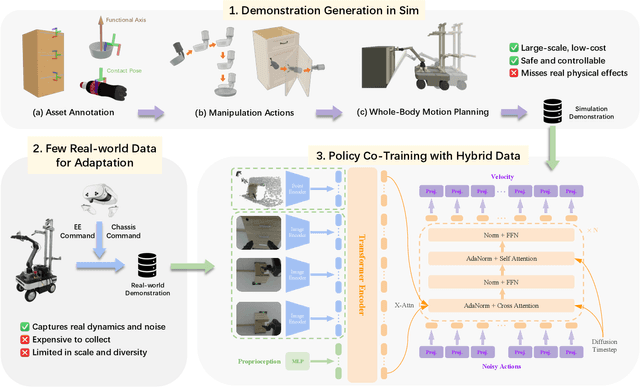
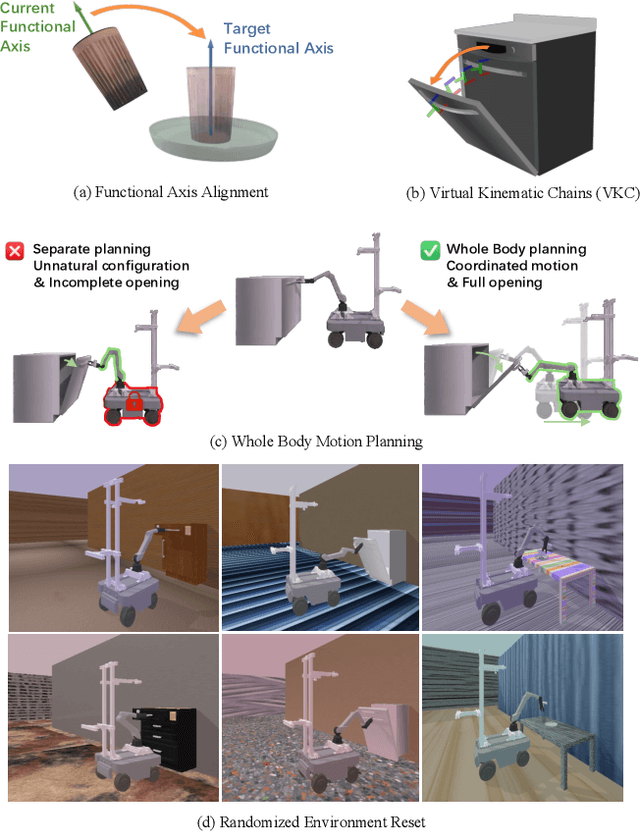
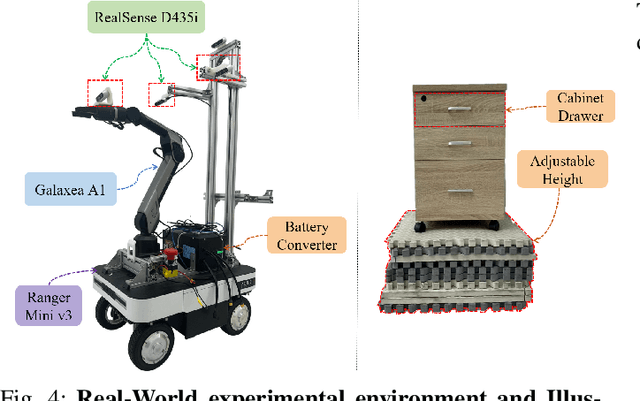
Abstract:Recent advances in robotics have been largely driven by imitation learning, which depends critically on large-scale, high-quality demonstration data. However, collecting such data remains a significant challenge-particularly for mobile manipulators, which must coordinate base locomotion and arm manipulation in high-dimensional, dynamic, and partially observable environments. Consequently, most existing research remains focused on simpler tabletop scenarios, leaving mobile manipulation relatively underexplored. To bridge this gap, we present \textit{MobRT}, a digital twin-based framework designed to simulate two primary categories of complex, whole-body tasks: interaction with articulated objects (e.g., opening doors and drawers) and mobile-base pick-and-place operations. \textit{MobRT} autonomously generates diverse and realistic demonstrations through the integration of virtual kinematic control and whole-body motion planning, enabling coherent and physically consistent execution. We evaluate the quality of \textit{MobRT}-generated data across multiple baseline algorithms, establishing a comprehensive benchmark and demonstrating a strong correlation between task success and the number of generated trajectories. Experiments integrating both simulated and real-world demonstrations confirm that our approach markedly improves policy generalization and performance, achieving robust results in both simulated and real-world environments.
Integrating Pathology Foundation Models and Spatial Transcriptomics for Cellular Decomposition from Histology Images
Jul 09, 2025Abstract:The rapid development of digital pathology and modern deep learning has facilitated the emergence of pathology foundation models that are expected to solve general pathology problems under various disease conditions in one unified model, with or without fine-tuning. In parallel, spatial transcriptomics has emerged as a transformative technology that enables the profiling of gene expression on hematoxylin and eosin (H&E) stained histology images. Spatial transcriptomics unlocks the unprecedented opportunity to dive into existing histology images at a more granular, cellular level. In this work, we propose a lightweight and training-efficient approach to predict cellular composition directly from H&E-stained histology images by leveraging information-enriched feature embeddings extracted from pre-trained pathology foundation models. By training a lightweight multi-layer perceptron (MLP) regressor on cell-type abundances derived via cell2location, our method efficiently distills knowledge from pathology foundation models and demonstrates the ability to accurately predict cell-type compositions from histology images, without physically performing the costly spatial transcriptomics. Our method demonstrates competitive performance compared to existing methods such as Hist2Cell, while significantly reducing computational complexity.
Diffusion Generative Modeling for Spatially Resolved Gene Expression Inference from Histology Images
Jan 26, 2025



Abstract:Spatial Transcriptomics (ST) allows a high-resolution measurement of RNA sequence abundance by systematically connecting cell morphology depicted in Hematoxylin and Eosin (H&E) stained histology images to spatially resolved gene expressions. ST is a time-consuming, expensive yet powerful experimental technique that provides new opportunities to understand cancer mechanisms at a fine-grained molecular level, which is critical for uncovering new approaches for disease diagnosis and treatments. Here, we present $\textbf{Stem}$ ($\textbf{S}$pa$\textbf{T}$ially resolved gene $\textbf{E}$xpression inference with diffusion $\textbf{M}$odel), a novel computational tool that leverages a conditional diffusion generative model to enable in silico gene expression inference from H&E stained images. Through better capturing the inherent stochasticity and heterogeneity in ST data, $\textbf{Stem}$ achieves state-of-the-art performance on spatial gene expression prediction and generates biologically meaningful gene profiles for new H&E stained images at test time. We evaluate the proposed algorithm on datasets with various tissue sources and sequencing platforms, where it demonstrates clear improvement over existing approaches. $\textbf{Stem}$ generates high-fidelity gene expression predictions that share similar gene variation levels as ground truth data, suggesting that our method preserves the underlying biological heterogeneity. Our proposed pipeline opens up the possibility of analyzing existing, easily accessible H&E stained histology images from a genomics point of view without physically performing gene expression profiling and empowers potential biological discovery from H&E stained histology images.
BASES: Large-scale Web Search User Simulation with Large Language Model based Agents
Feb 27, 2024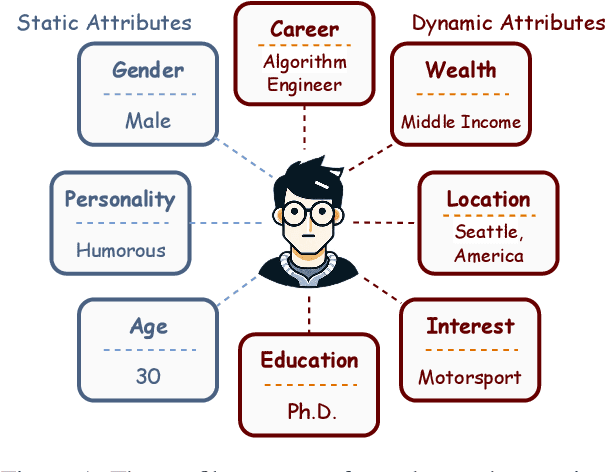
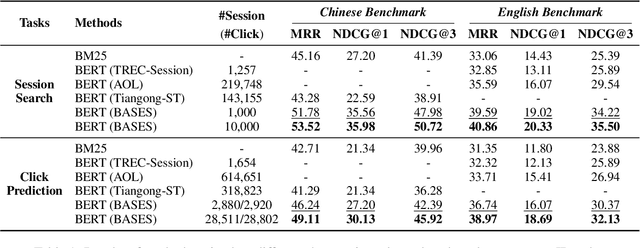
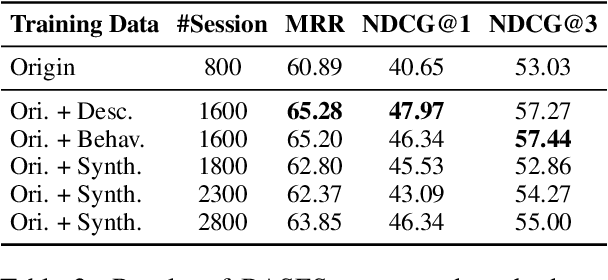
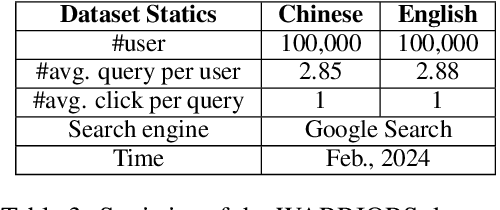
Abstract:Due to the excellent capacities of large language models (LLMs), it becomes feasible to develop LLM-based agents for reliable user simulation. Considering the scarcity and limit (e.g., privacy issues) of real user data, in this paper, we conduct large-scale user simulation for web search, to improve the analysis and modeling of user search behavior. Specially, we propose BASES, a novel user simulation framework with LLM-based agents, designed to facilitate comprehensive simulations of web search user behaviors. Our simulation framework can generate unique user profiles at scale, which subsequently leads to diverse search behaviors. To demonstrate the effectiveness of BASES, we conduct evaluation experiments based on two human benchmarks in both Chinese and English, demonstrating that BASES can effectively simulate large-scale human-like search behaviors. To further accommodate the research on web search, we develop WARRIORS, a new large-scale dataset encompassing web search user behaviors, including both Chinese and English versions, which can greatly bolster research in the field of information retrieval. Our code and data will be publicly released soon.
Cell Spatial Analysis in Crohn's Disease: Unveiling Local Cell Arrangement Pattern with Graph-based Signatures
Aug 20, 2023Abstract:Crohn's disease (CD) is a chronic and relapsing inflammatory condition that affects segments of the gastrointestinal tract. CD activity is determined by histological findings, particularly the density of neutrophils observed on Hematoxylin and Eosin stains (H&E) imaging. However, understanding the broader morphometry and local cell arrangement beyond cell counting and tissue morphology remains challenging. To address this, we characterize six distinct cell types from H&E images and develop a novel approach for the local spatial signature of each cell. Specifically, we create a 10-cell neighborhood matrix, representing neighboring cell arrangements for each individual cell. Utilizing t-SNE for non-linear spatial projection in scatter-plot and Kernel Density Estimation contour-plot formats, our study examines patterns of differences in the cellular environment associated with the odds ratio of spatial patterns between active CD and control groups. This analysis is based on data collected at the two research institutes. The findings reveal heterogeneous nearest-neighbor patterns, signifying distinct tendencies of cell clustering, with a particular focus on the rectum region. These variations underscore the impact of data heterogeneity on cell spatial arrangements in CD patients. Moreover, the spatial distribution disparities between the two research sites highlight the significance of collaborative efforts among healthcare organizations. All research analysis pipeline tools are available at https://github.com/MASILab/cellNN.
Polycraft World AI Lab : An Extensible Platform for Evaluating Artificial Intelligence Agents
Jan 27, 2023Abstract:As artificial intelligence research advances, the platforms used to evaluate AI agents need to adapt and grow to continue to challenge them. We present the Polycraft World AI Lab (PAL), a task simulator with an API based on the Minecraft mod Polycraft World. Our platform is built to allow AI agents with different architectures to easily interact with the Minecraft world, train and be evaluated in multiple tasks. PAL enables the creation of tasks in a flexible manner as well as having the capability to manipulate any aspect of the task during an evaluation. All actions taken by AI agents and external actors (non-player-characters, NPCs) in the open-world environment are logged to streamline evaluation. Here we present two custom tasks on the PAL platform, one focused on multi-step planning and one focused on navigation, and evaluations of agents solving them. In summary, we report a versatile and extensible AI evaluation platform with a low barrier to entry for AI researchers to utilize.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge