Noriaki Hashimoto
Explainable Classifier for Malignant Lymphoma Subtyping via Cell Graph and Image Fusion
Mar 02, 2025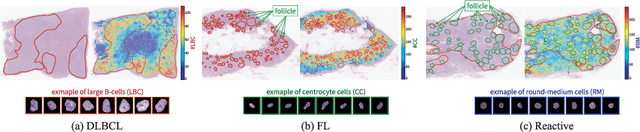
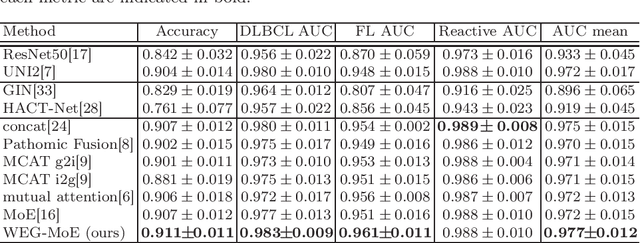
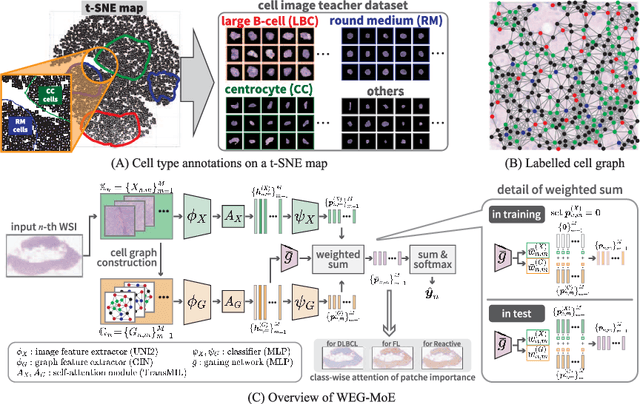
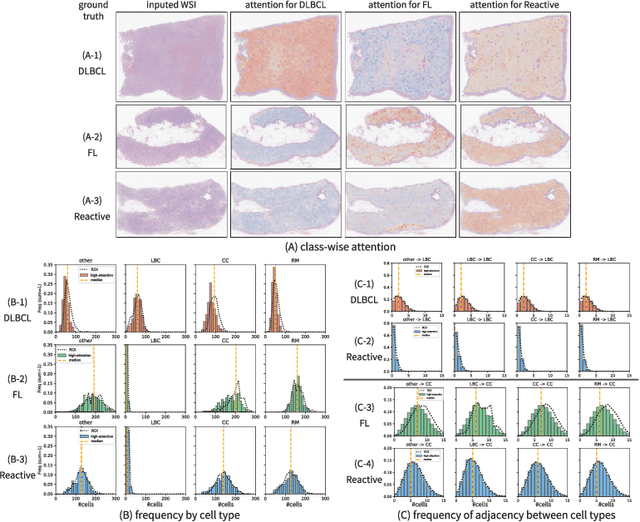
Abstract:Malignant lymphoma subtype classification directly impacts treatment strategies and patient outcomes, necessitating classification models that achieve both high accuracy and sufficient explainability. This study proposes a novel explainable Multi-Instance Learning (MIL) framework that identifies subtype-specific Regions of Interest (ROIs) from Whole Slide Images (WSIs) while integrating cell distribution characteristics and image information. Our framework simultaneously addresses three objectives: (1) indicating appropriate ROIs for each subtype, (2) explaining the frequency and spatial distribution of characteristic cell types, and (3) achieving high-accuracy subtyping by leveraging both image and cell-distribution modalities. The proposed method fuses cell graph and image features extracted from each patch in the WSI using a Mixture-of-Experts (MoE) approach and classifies subtypes within an MIL framework. Experiments on a dataset of 1,233 WSIs demonstrate that our approach achieves state-of-the-art accuracy among ten comparative methods and provides region-level and cell-level explanations that align with a pathologist's perspectives.
Distributionally Robust Active Learning for Gaussian Process Regression
Feb 24, 2025Abstract:Gaussian process regression (GPR) or kernel ridge regression is a widely used and powerful tool for nonlinear prediction. Therefore, active learning (AL) for GPR, which actively collects data labels to achieve an accurate prediction with fewer data labels, is an important problem. However, existing AL methods do not theoretically guarantee prediction accuracy for target distribution. Furthermore, as discussed in the distributionally robust learning literature, specifying the target distribution is often difficult. Thus, this paper proposes two AL methods that effectively reduce the worst-case expected error for GPR, which is the worst-case expectation in target distribution candidates. We show an upper bound of the worst-case expected squared error, which suggests that the error will be arbitrarily small by a finite number of data labels under mild conditions. Finally, we demonstrate the effectiveness of the proposed methods through synthetic and real-world datasets.
Generalized Kernel Inducing Points by Duality Gap for Dataset Distillation
Feb 18, 2025Abstract:We propose Duality Gap KIP (DGKIP), an extension of the Kernel Inducing Points (KIP) method for dataset distillation. While existing dataset distillation methods often rely on bi-level optimization, DGKIP eliminates the need for such optimization by leveraging duality theory in convex programming. The KIP method has been introduced as a way to avoid bi-level optimization; however, it is limited to the squared loss and does not support other loss functions (e.g., cross-entropy or hinge loss) that are more suitable for classification tasks. DGKIP addresses this limitation by exploiting an upper bound on parameter changes after dataset distillation using the duality gap, enabling its application to a wider range of loss functions. We also characterize theoretical properties of DGKIP by providing upper bounds on the test error and prediction consistency after dataset distillation. Experimental results on standard benchmarks such as MNIST and CIFAR-10 demonstrate that DGKIP retains the efficiency of KIP while offering broader applicability and robust performance.
Distributionally Robust Coreset Selection under Covariate Shift
Jan 24, 2025Abstract:Coreset selection, which involves selecting a small subset from an existing training dataset, is an approach to reducing training data, and various approaches have been proposed for this method. In practical situations where these methods are employed, it is often the case that the data distributions differ between the development phase and the deployment phase, with the latter being unknown. Thus, it is challenging to select an effective subset of training data that performs well across all deployment scenarios. We therefore propose Distributionally Robust Coreset Selection (DRCS). DRCS theoretically derives an estimate of the upper bound for the worst-case test error, assuming that the future covariate distribution may deviate within a defined range from the training distribution. Furthermore, by selecting instances in a way that suppresses the estimate of the upper bound for the worst-case test error, DRCS achieves distributionally robust training instance selection. This study is primarily applicable to convex training computation, but we demonstrate that it can also be applied to deep learning under appropriate approximations. In this paper, we focus on covariate shift, a type of data distribution shift, and demonstrate the effectiveness of DRCS through experiments.
Distributionally Robust Safe Sample Screening
Jun 10, 2024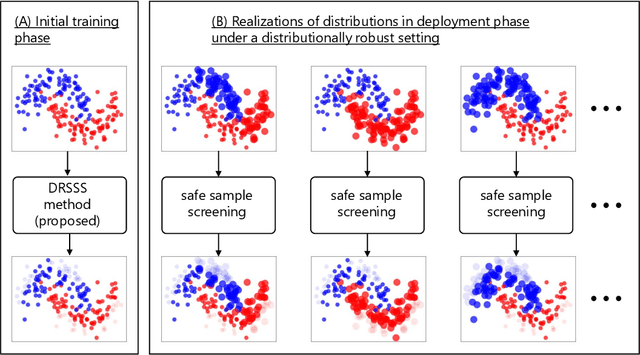

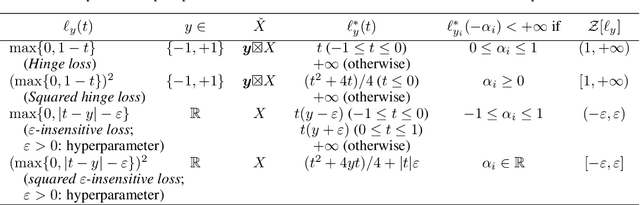
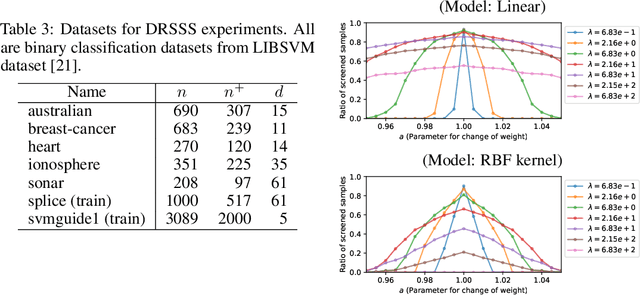
Abstract:In this study, we propose a machine learning method called Distributionally Robust Safe Sample Screening (DRSSS). DRSSS aims to identify unnecessary training samples, even when the distribution of the training samples changes in the future. To achieve this, we effectively combine the distributionally robust (DR) paradigm, which aims to enhance model robustness against variations in data distribution, with the safe sample screening (SSS), which identifies unnecessary training samples prior to model training. Since we need to consider an infinite number of scenarios regarding changes in the distribution, we applied SSS because it does not require model training after the change of the distribution. In this paper, we employed the covariate shift framework to represent the distribution of training samples and reformulated the DR covariate-shift problem as a weighted empirical risk minimization problem, where the weights are subject to uncertainty within a predetermined range. By extending the existing SSS technique to accommodate this weight uncertainty, the DRSSS method is capable of reliably identifying unnecessary samples under any future distribution within a specified range. We provide a theoretical guarantee for the DRSSS method and validate its performance through numerical experiments on both synthetic and real-world datasets.
Distributionally Robust Safe Screening
Apr 25, 2024Abstract:In this study, we propose a method Distributionally Robust Safe Screening (DRSS), for identifying unnecessary samples and features within a DR covariate shift setting. This method effectively combines DR learning, a paradigm aimed at enhancing model robustness against variations in data distribution, with safe screening (SS), a sparse optimization technique designed to identify irrelevant samples and features prior to model training. The core concept of the DRSS method involves reformulating the DR covariate-shift problem as a weighted empirical risk minimization problem, where the weights are subject to uncertainty within a predetermined range. By extending the SS technique to accommodate this weight uncertainty, the DRSS method is capable of reliably identifying unnecessary samples and features under any future distribution within a specified range. We provide a theoretical guarantee of the DRSS method and validate its performance through numerical experiments on both synthetic and real-world datasets.
Mixing Histopathology Prototypes into Robust Slide-Level Representations for Cancer Subtyping
Oct 19, 2023Abstract:Whole-slide image analysis via the means of computational pathology often relies on processing tessellated gigapixel images with only slide-level labels available. Applying multiple instance learning-based methods or transformer models is computationally expensive as, for each image, all instances have to be processed simultaneously. The MLP-Mixer is an under-explored alternative model to common vision transformers, especially for large-scale datasets. Due to the lack of a self-attention mechanism, they have linear computational complexity to the number of input patches but achieve comparable performance on natural image datasets. We propose a combination of feature embedding and clustering to preprocess the full whole-slide image into a reduced prototype representation which can then serve as input to a suitable MLP-Mixer architecture. Our experiments on two public benchmarks and one inhouse malignant lymphoma dataset show comparable performance to current state-of-the-art methods, while achieving lower training costs in terms of computational time and memory load. Code is publicly available at https://github.com/butkej/ProtoMixer.
* The final authenticated publication is available online at https://doi.org/10.1007/978-3-031-45676-3_12
Generalized Low-Rank Update: Model Parameter Bounds for Low-Rank Training Data Modifications
Jun 22, 2023Abstract:In this study, we have developed an incremental machine learning (ML) method that efficiently obtains the optimal model when a small number of instances or features are added or removed. This problem holds practical importance in model selection, such as cross-validation (CV) and feature selection. Among the class of ML methods known as linear estimators, there exists an efficient model update framework called the low-rank update that can effectively handle changes in a small number of rows and columns within the data matrix. However, for ML methods beyond linear estimators, there is currently no comprehensive framework available to obtain knowledge about the updated solution within a specific computational complexity. In light of this, our study introduces a method called the Generalized Low-Rank Update (GLRU) which extends the low-rank update framework of linear estimators to ML methods formulated as a certain class of regularized empirical risk minimization, including commonly used methods such as SVM and logistic regression. The proposed GLRU method not only expands the range of its applicability but also provides information about the updated solutions with a computational complexity proportional to the amount of dataset changes. To demonstrate the effectiveness of the GLRU method, we conduct experiments showcasing its efficiency in performing cross-validation and feature selection compared to other baseline methods.
Transformer-based Personalized Attention Mechanism (PersAM) for Medical Images with Clinical Records
Jun 07, 2022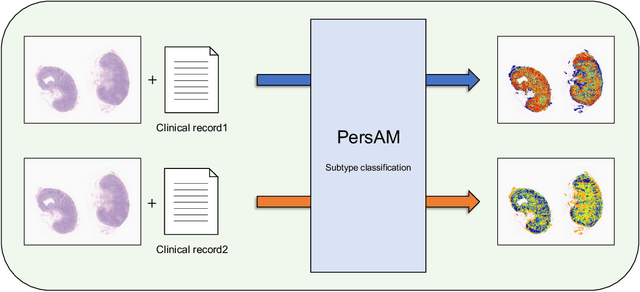
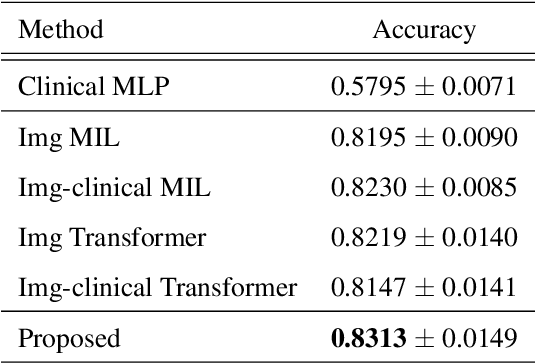
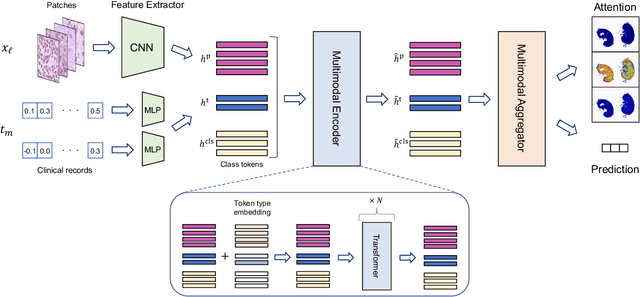
Abstract:In medical image diagnosis, identifying the attention region, i.e., the region of interest for which the diagnosis is made, is an important task. Various methods have been developed to automatically identify target regions from given medical images. However, in actual medical practice, the diagnosis is made based not only on the images but also on a variety of clinical records. This means that pathologists examine medical images with some prior knowledge of the patients and that the attention regions may change depending on the clinical records. In this study, we propose a method called the Personalized Attention Mechanism (PersAM), by which the attention regions in medical images are adaptively changed according to the clinical records. The primary idea of the PersAM method is to encode the relationships between the medical images and clinical records using a variant of Transformer architecture. To demonstrate the effectiveness of the PersAM method, we applied it to a large-scale digital pathology problem of identifying the subtypes of 842 malignant lymphoma patients based on their gigapixel whole slide images and clinical records.
Case-based similar image retrieval for weakly annotated large histopathological images of malignant lymphoma using deep metric learning
Jul 09, 2021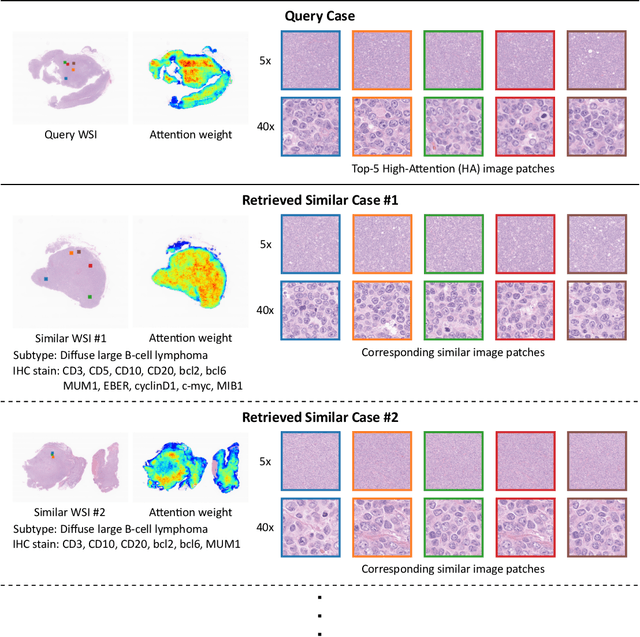
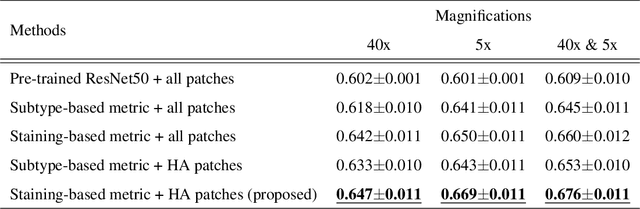
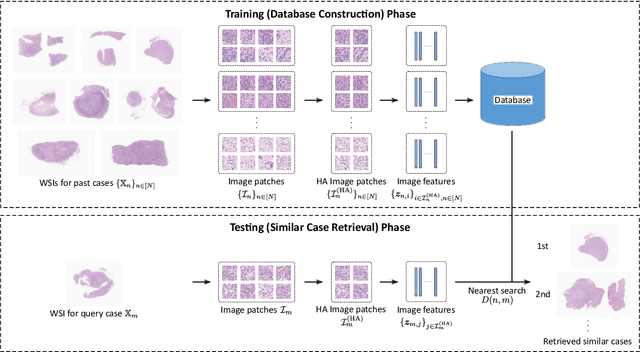
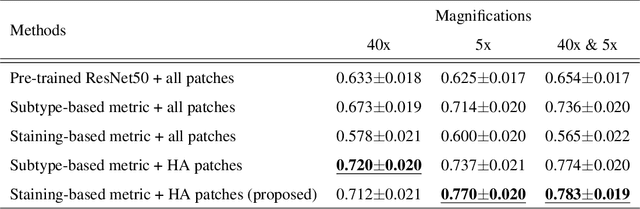
Abstract:In the present study, we propose a novel case-based similar image retrieval (SIR) method for hematoxylin and eosin (H&E)-stained histopathological images of malignant lymphoma. When a whole slide image (WSI) is used as an input query, it is desirable to be able to retrieve similar cases by focusing on image patches in pathologically important regions such as tumor cells. To address this problem, we employ attention-based multiple instance learning, which enables us to focus on tumor-specific regions when the similarity between cases is computed. Moreover, we employ contrastive distance metric learning to incorporate immunohistochemical (IHC) staining patterns as useful supervised information for defining appropriate similarity between heterogeneous malignant lymphoma cases. In the experiment with 249 malignant lymphoma patients, we confirmed that the proposed method exhibited higher evaluation measures than the baseline case-based SIR methods. Furthermore, the subjective evaluation by pathologists revealed that our similarity measure using IHC staining patterns is appropriate for representing the similarity of H&E-stained tissue images for malignant lymphoma.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge