Yusuke Takagi
Transformer-based Personalized Attention Mechanism (PersAM) for Medical Images with Clinical Records
Jun 07, 2022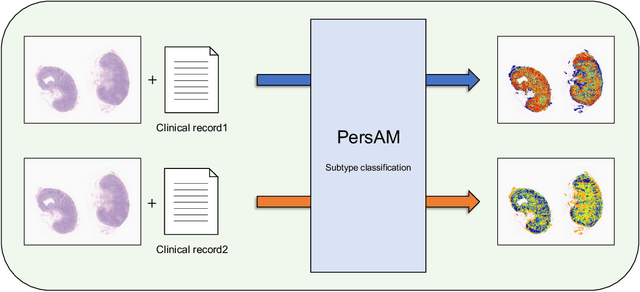
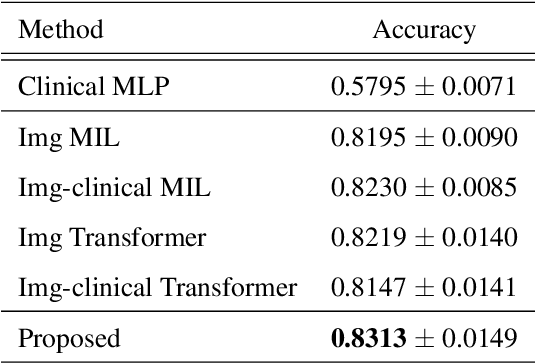
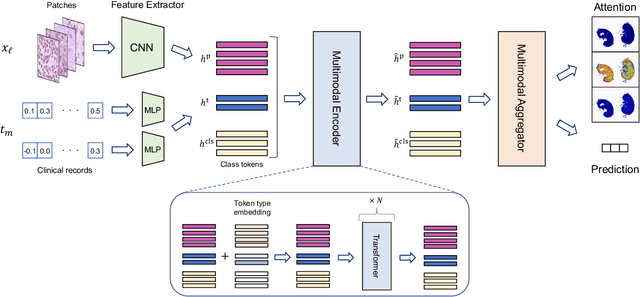
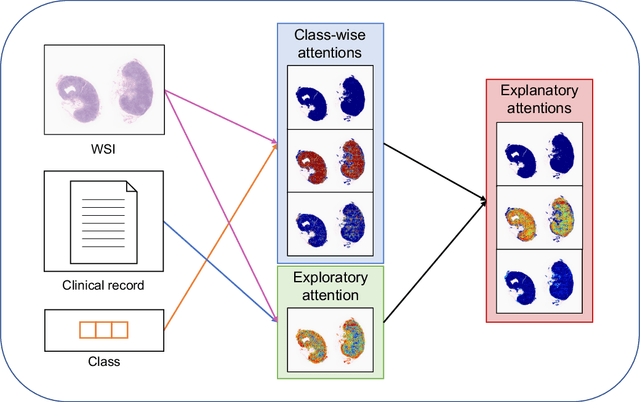
Abstract:In medical image diagnosis, identifying the attention region, i.e., the region of interest for which the diagnosis is made, is an important task. Various methods have been developed to automatically identify target regions from given medical images. However, in actual medical practice, the diagnosis is made based not only on the images but also on a variety of clinical records. This means that pathologists examine medical images with some prior knowledge of the patients and that the attention regions may change depending on the clinical records. In this study, we propose a method called the Personalized Attention Mechanism (PersAM), by which the attention regions in medical images are adaptively changed according to the clinical records. The primary idea of the PersAM method is to encode the relationships between the medical images and clinical records using a variant of Transformer architecture. To demonstrate the effectiveness of the PersAM method, we applied it to a large-scale digital pathology problem of identifying the subtypes of 842 malignant lymphoma patients based on their gigapixel whole slide images and clinical records.
Case-based similar image retrieval for weakly annotated large histopathological images of malignant lymphoma using deep metric learning
Jul 09, 2021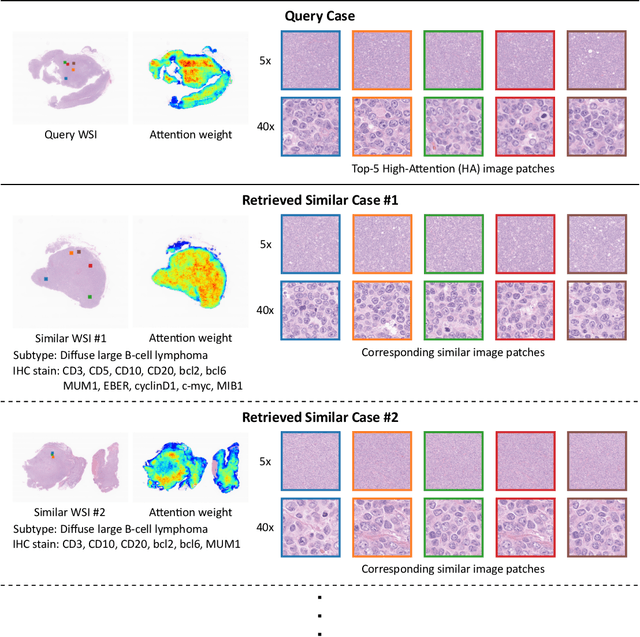
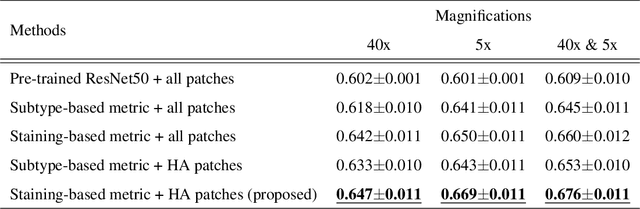
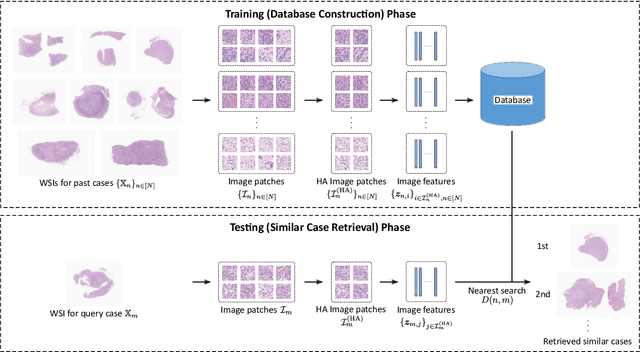
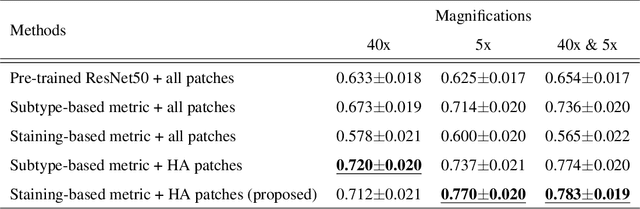
Abstract:In the present study, we propose a novel case-based similar image retrieval (SIR) method for hematoxylin and eosin (H&E)-stained histopathological images of malignant lymphoma. When a whole slide image (WSI) is used as an input query, it is desirable to be able to retrieve similar cases by focusing on image patches in pathologically important regions such as tumor cells. To address this problem, we employ attention-based multiple instance learning, which enables us to focus on tumor-specific regions when the similarity between cases is computed. Moreover, we employ contrastive distance metric learning to incorporate immunohistochemical (IHC) staining patterns as useful supervised information for defining appropriate similarity between heterogeneous malignant lymphoma cases. In the experiment with 249 malignant lymphoma patients, we confirmed that the proposed method exhibited higher evaluation measures than the baseline case-based SIR methods. Furthermore, the subjective evaluation by pathologists revealed that our similarity measure using IHC staining patterns is appropriate for representing the similarity of H&E-stained tissue images for malignant lymphoma.
Multi-scale domain-adversarial multiple-instance CNN for cancer subtype classification with non-annotated histopathological images
Jan 06, 2020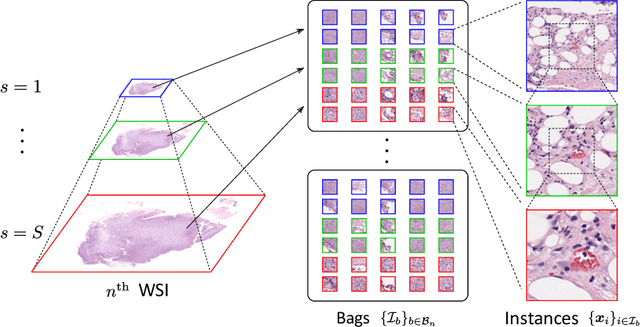
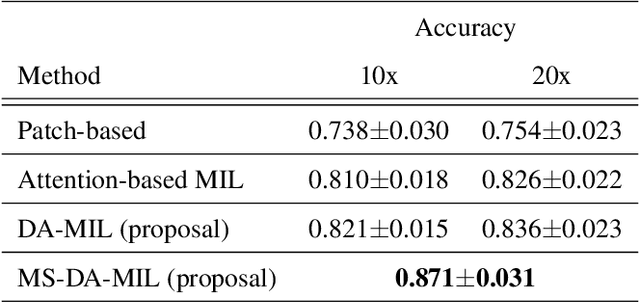
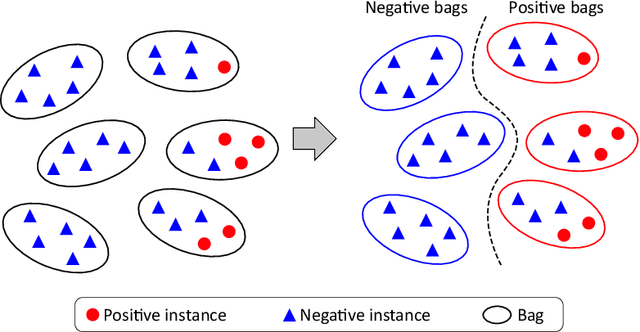
Abstract:We propose a new method for cancer subtype classification from histopathological images, which can automatically detect tumor-specific features in a given whole slide image (WSI). The cancer subtype should be classified by referring to a WSI, i.e., a large size image (typically 40,000x40,000 pixels) of an entire pathological tissue slide, which consists of cancer and non-cancer portions. One difficulty for constructing cancer subtype classifiers comes from the high cost needed for annotating WSIs; without annotation, we have to construct the tumor region detector without knowing true labels. Furthermore, both global and local image features must be extracted from the WSI by changing the magnifications of the image. In addition, the image features should be stably detected against the variety/difference of staining among the hospitals/specimen. In this paper, we develop a new CNN-based cancer subtype classification method by effectively combining multiple-instance, domain adversarial, and multi-scale learning frameworks that can overcome these practical difficulties. When the proposed method was applied to malignant lymphoma subtype classifications of 196 cases collected from multiple hospitals, the classification performance was significantly better than the standard CNN or other conventional methods, and the accuracy was favorably compared to that of standard pathologists. In addition, we confirmed by immunostaining and expert pathologist's visual inspections that the tumor regions were correctly detected.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge