Nina Lukashina
Structure-Aware E(3)-Invariant Molecular Conformer Aggregation Networks
Feb 03, 2024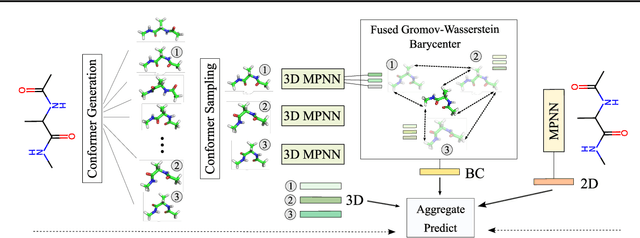

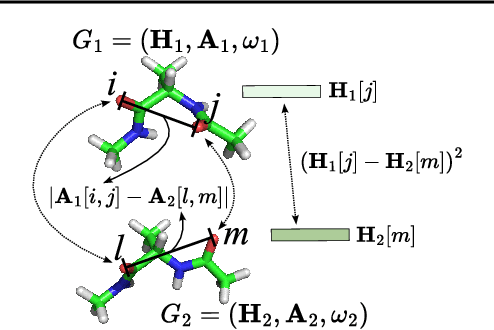

Abstract:A molecule's 2D representation consists of its atoms, their attributes, and the molecule's covalent bonds. A 3D (geometric) representation of a molecule is called a conformer and consists of its atom types and Cartesian coordinates. Every conformer has a potential energy, and the lower this energy, the more likely it occurs in nature. Most existing machine learning methods for molecular property prediction consider either 2D molecular graphs or 3D conformer structure representations in isolation. Inspired by recent work on using ensembles of conformers in conjunction with 2D graph representations, we propose E(3)-invariant molecular conformer aggregation networks. The method integrates a molecule's 2D representation with that of multiple of its conformers. Contrary to prior work, we propose a novel 2D--3D aggregation mechanism based on a differentiable solver for the \emph{Fused Gromov-Wasserstein Barycenter} problem and the use of an efficient online conformer generation method based on distance geometry. We show that the proposed aggregation mechanism is E(3) invariant and provides an efficient GPU implementation. Moreover, we demonstrate that the aggregation mechanism helps to outperform state-of-the-art property prediction methods on established datasets significantly.
Traffic4cast at NeurIPS 2021 -- Temporal and Spatial Few-Shot Transfer Learning in Gridded Geo-Spatial Processes
Apr 01, 2022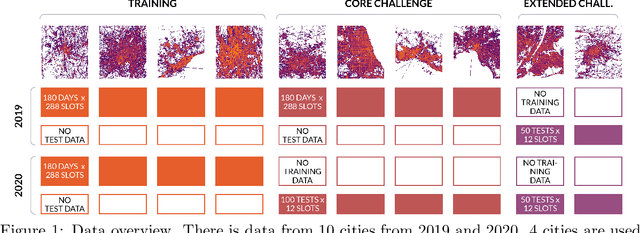
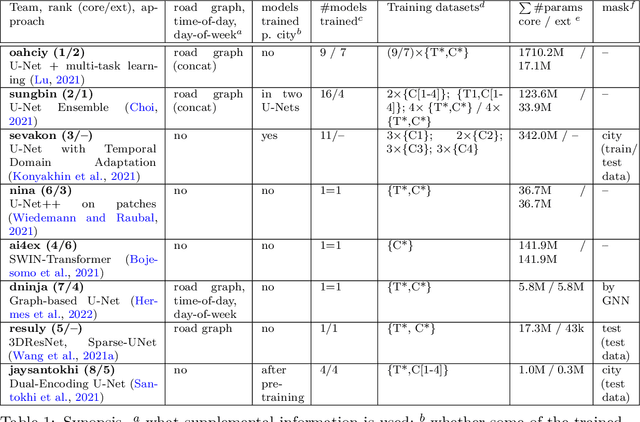
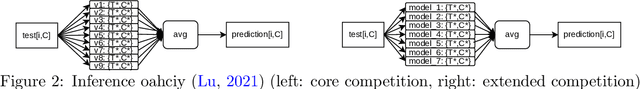
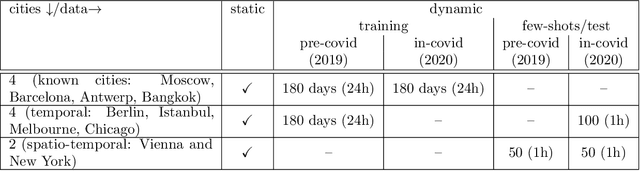
Abstract:The IARAI Traffic4cast competitions at NeurIPS 2019 and 2020 showed that neural networks can successfully predict future traffic conditions 1 hour into the future on simply aggregated GPS probe data in time and space bins. We thus reinterpreted the challenge of forecasting traffic conditions as a movie completion task. U-Nets proved to be the winning architecture, demonstrating an ability to extract relevant features in this complex real-world geo-spatial process. Building on the previous competitions, Traffic4cast 2021 now focuses on the question of model robustness and generalizability across time and space. Moving from one city to an entirely different city, or moving from pre-COVID times to times after COVID hit the world thus introduces a clear domain shift. We thus, for the first time, release data featuring such domain shifts. The competition now covers ten cities over 2 years, providing data compiled from over 10^12 GPS probe data. Winning solutions captured traffic dynamics sufficiently well to even cope with these complex domain shifts. Surprisingly, this seemed to require only the previous 1h traffic dynamic history and static road graph as input.
Solving Traffic4Cast Competition with U-Net and Temporal Domain Adaptation
Nov 05, 2021
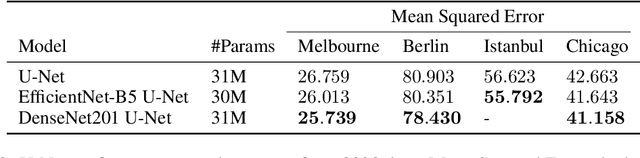


Abstract:In this technical report, we present our solution to the Traffic4Cast 2021 Core Challenge, in which participants were asked to develop algorithms for predicting a traffic state 60 minutes ahead, based on the information from the previous hour, in 4 different cities. In contrast to the previously held competitions, this year's challenge focuses on the temporal domain shift in traffic due to the COVID-19 pandemic. Following the past success of U-Net, we utilize it for predicting future traffic maps. Additionally, we explore the usage of pre-trained encoders such as DenseNet and EfficientNet and employ multiple domain adaptation techniques to fight the domain shift. Our solution has ranked third in the final competition. The code is available at https://github.com/jbr-ai-labs/traffic4cast-2021.
Lipophilicity Prediction with Multitask Learning and Molecular Substructures Representation
Nov 24, 2020


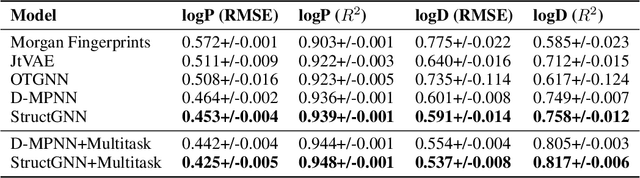
Abstract:Lipophilicity is one of the factors determining the permeability of the cell membrane to a drug molecule. Hence, accurate lipophilicity prediction is an essential step in the development of new drugs. In this paper, we introduce a novel approach to encoding additional graph information by extracting molecular substructures. By adding a set of generalized atomic features of these substructures to an established Direct Message Passing Neural Network (D-MPNN) we were able to achieve a new state-of-the-art result at the task of prediction of two main lipophilicity coefficients, namely logP and logD descriptors. We further improve our approach by employing a multitask approach to predict logP and logD values simultaneously. Additionally, we present a study of the model performance on symmetric and asymmetric molecules, that may yield insight for further research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge