Mohammad Eslami
Federated Learning for Diabetic Retinopathy Diagnosis: Enhancing Accuracy and Generalizability in Under-Resourced Regions
Oct 30, 2024Abstract:Diabetic retinopathy is the leading cause of vision loss in working-age adults worldwide, yet under-resourced regions lack ophthalmologists. Current state-of-the-art deep learning systems struggle at these institutions due to limited generalizability. This paper explores a novel federated learning system for diabetic retinopathy diagnosis with the EfficientNetB0 architecture to leverage fundus data from multiple institutions to improve diagnostic generalizability at under-resourced hospitals while preserving patient-privacy. The federated model achieved 93.21% accuracy in five-category classification on an unseen dataset and 91.05% on lower-quality images from a simulated under-resourced institution. The model was deployed onto two apps for quick and accurate diagnosis.
Affective Medical Estimation and Decision Making via Visualized Learning and Deep Learning
May 09, 2022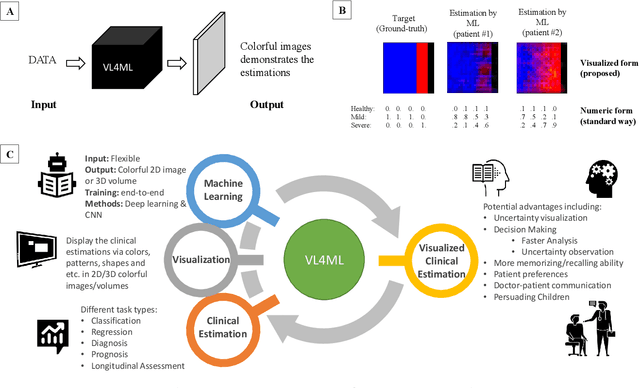
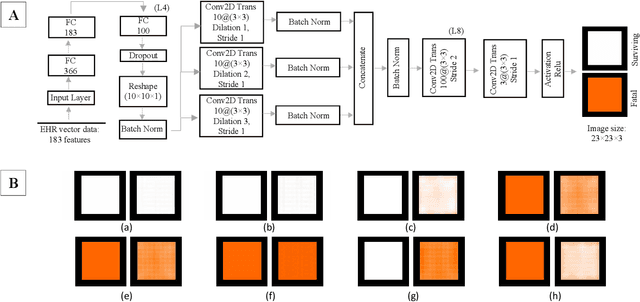
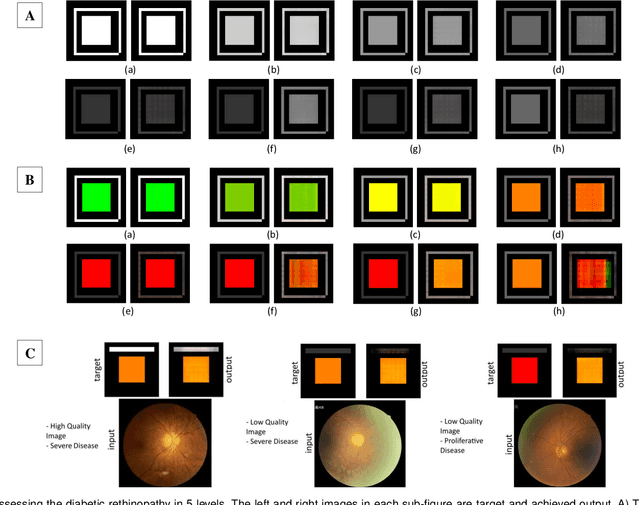
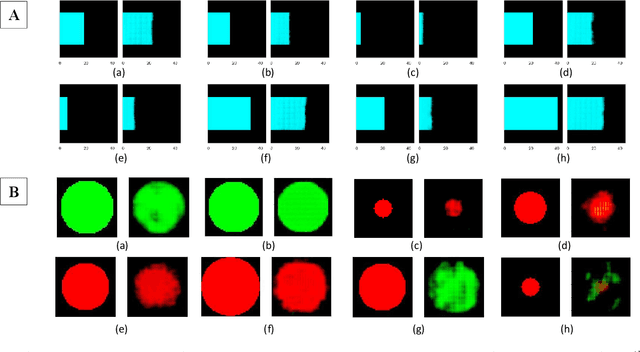
Abstract:With the advent of sophisticated machine learning (ML) techniques and the promising results they yield, especially in medical applications, where they have been investigated for different tasks to enhance the decision-making process. Since visualization is such an effective tool for human comprehension, memorization, and judgment, we have presented a first-of-its-kind estimation approach we refer to as Visualized Learning for Machine Learning (VL4ML) that not only can serve to assist physicians and clinicians in making reasoned medical decisions, but it also allows to appreciate the uncertainty visualization, which could raise incertitude in making the appropriate classification or prediction. For the proof of concept, and to demonstrate the generalized nature of this visualized estimation approach, five different case studies are examined for different types of tasks including classification, regression, and longitudinal prediction. A survey analysis with more than 100 individuals is also conducted to assess users' feedback on this visualized estimation method. The experiments and the survey demonstrate the practical merits of the VL4ML that include: (1) appreciating visually clinical/medical estimations; (2) getting closer to the patients' preferences; (3) improving doctor-patient communication, and (4) visualizing the uncertainty introduced through the black box effect of the deployed ML algorithm. All the source codes are shared via a GitHub repository.
Deep Variational Clustering Framework for Self-labeling of Large-scale Medical Images
Sep 22, 2021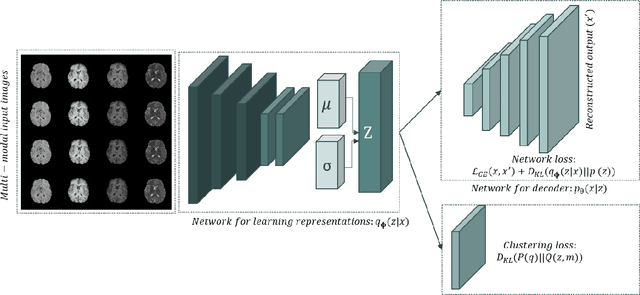
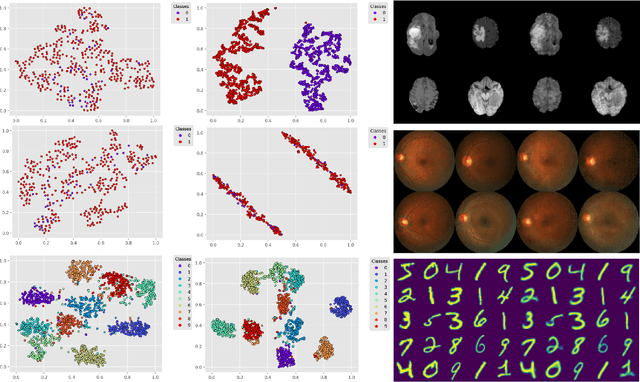
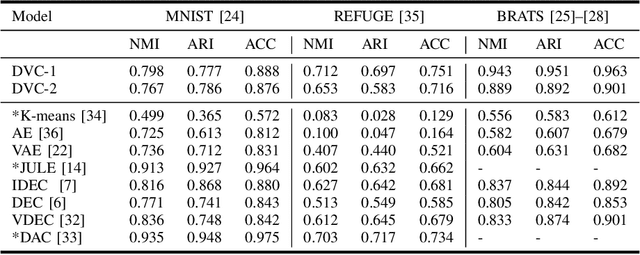
Abstract:We propose a Deep Variational Clustering (DVC) framework for unsupervised representation learning and clustering of large-scale medical images. DVC simultaneously learns the multivariate Gaussian posterior through the probabilistic convolutional encoder and the likelihood distribution with the probabilistic convolutional decoder; and optimizes cluster labels assignment. Here, the learned multivariate Gaussian posterior captures the latent distribution of a large set of unlabeled images. Then, we perform unsupervised clustering on top of the variational latent space using a clustering loss. In this approach, the probabilistic decoder helps to prevent the distortion of data points in the latent space and to preserve the local structure of data generating distribution. The training process can be considered as a self-training process to refine the latent space and simultaneously optimizing cluster assignments iteratively. We evaluated our proposed framework on three public datasets that represented different medical imaging modalities. Our experimental results show that our proposed framework generalizes better across different datasets. It achieves compelling results on several medical imaging benchmarks. Thus, our approach offers potential advantages over conventional deep unsupervised learning in real-world applications. The source code of the method and all the experiments are available publicly at: https://github.com/csfarzin/DVC
Feasibility Assessment of Multitasking in MRI Neuroimaging Analysis: Tissue Segmentation, Cross-Modality Conversion and Bias correction
May 31, 2021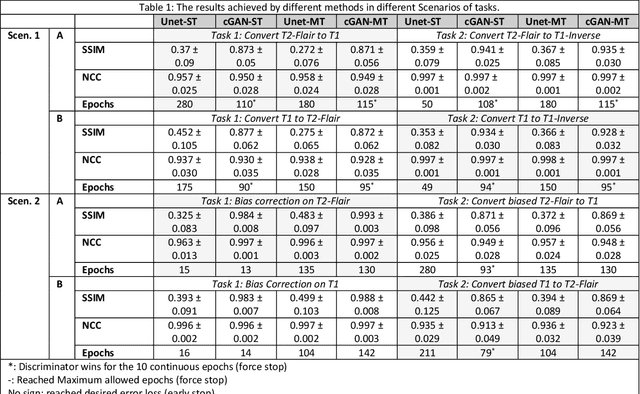
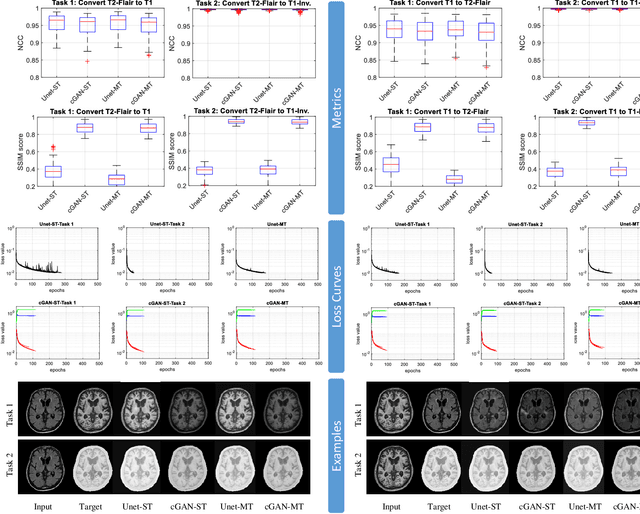
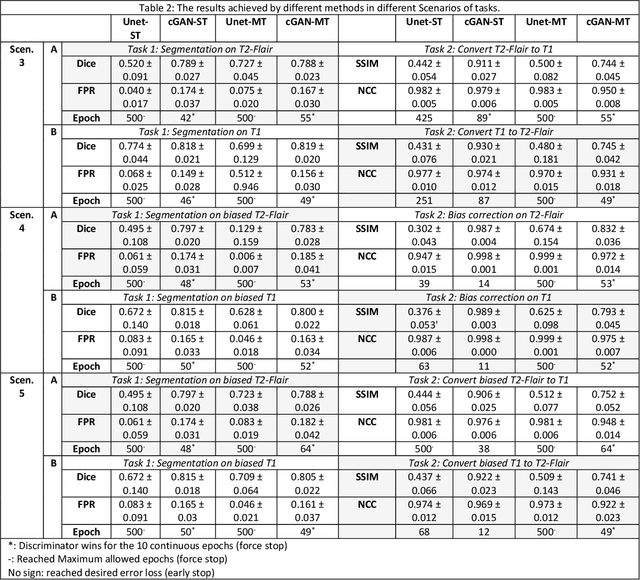
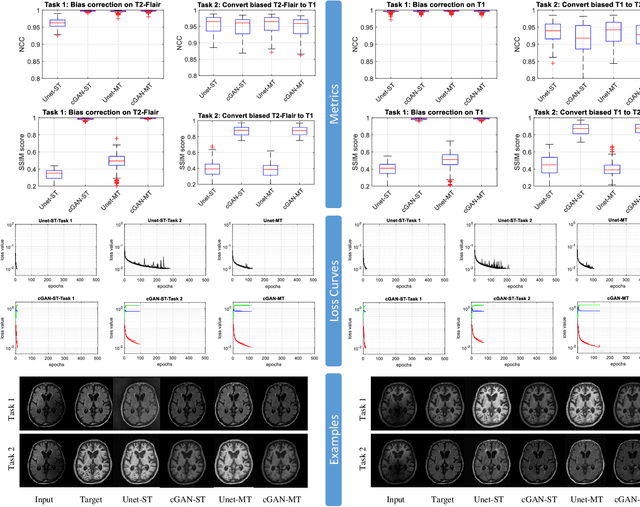
Abstract:Neuroimaging is essential in brain studies for the diagnosis and identification of disease, structure, and function of the brain in its healthy and disease states. Literature shows that there are advantages of multitasking with some deep learning (DL) schemes in challenging neuroimaging applications. This study examines the feasibility of using multitasking in three different applications, including tissue segmentation, cross-modality conversion, and bias-field correction. These applications reflect five different scenarios in which multitasking is explored and 280 training and testing sessions conducted for empirical evaluations. Two well-known networks, U-Net as a well-known convolutional neural network architecture, and a closed architecture based on the conditional generative adversarial network are implemented. Different metrics such as the normalized cross-correlation coefficient and Dice scores are used for comparison of methods and results of the different experiments. Statistical analysis is also provided by paired t-test. The present study explores the pros and cons of these methods and their practical impacts on multitasking in different implementation scenarios. This investigation shows that bias correction and cross-modality conversion applications are significantly easier than the segmentation application, and having multitasking with segmentation is not reasonable if one of them is identified as the main target application. However, when the main application is the segmentation of tissues, multitasking with cross-modality conversion is beneficial, especially for the U-net architecture.
SignCol: Open-Source Software for Collecting Sign Language Gestures
Oct 31, 2019



Abstract:Sign(ed) languages use gestures, such as hand or head movements, for communication. Sign language recognition is an assistive technology for individuals with hearing disability and its goal is to improve such individuals' life quality by facilitating their social involvement. Since sign languages are vastly varied in alphabets, as known as signs, a sign recognition software should be capable of handling eight different types of sign combinations, e.g. numbers, letters, words and sentences. Due to the intrinsic complexity and diversity of symbolic gestures, recognition algorithms need a comprehensive visual dataset to learn by. In this paper, we describe the design and implementation of a Microsoft Kinect-based open source software, called SignCol, for capturing and saving the gestures used in sign languages. Our work supports a multi-language database and reports the recorded items statistics. SignCol can capture and store colored(RGB) frames, depth frames, infrared frames, body index frames, coordinate mapped color-body frames, skeleton information of each frame and camera parameters simultaneously.
Automatic vocal tract landmark localization from midsagittal MRI data
Jul 18, 2019



Abstract:The various speech sounds of a language are obtained by varying the shape and position of the articulators surrounding the vocal tract. Analyzing their variability is crucial for understanding speech production, diagnosing speech and swallowing disorders and building intuitive applications for rehabilitation. Magnetic Resonance Imaging (MRI) is currently the most harmless powerful imaging modality used for this purpose. Identifying key anatomical landmarks on it is a pre-requisite for further analyses. This is a challenging task considering the high inter- and intra-speaker variability and the mutual interaction between the articulators. This study intends to solve this issue automatically for the first time. For this purpose, midsagittal anatomical MRI for 9 speakers sustaining 62 articulations and annotated with the location of 21 key anatomical landmarks are considered. Four state-of-the-art methods, including deep learning methods, are adapted from the literature for facial landmark localization and human pose estimation and evaluated. Furthermore, an approach based on the description of each landmark location as a heat-map image stored in a channel of a single multi-channel image embedding all landmarks is proposed. The generation of such a multi-channel image from an input MRI image is tested through two deep learning networks, one taken from the literature and one designed on purpose in this study, the flat-net. Results show that the flat-net approach outperforms the other methods, leading to an overall Root Mean Square Error of 3.4~pixels/0.34~cm obtained in a leave-one-out procedure over the speakers. All of the codes are publicly available on GitHub.
Image to Images Translation for Multi-Task Organ Segmentation and Bone Suppression in Chest X-Ray Radiography
Jun 24, 2019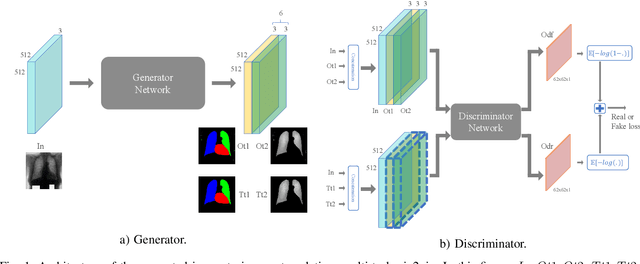
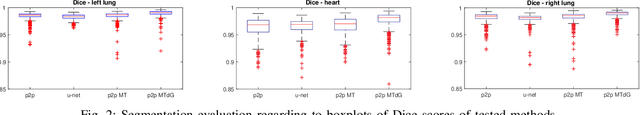
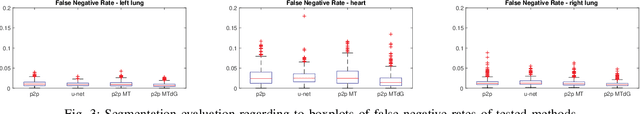
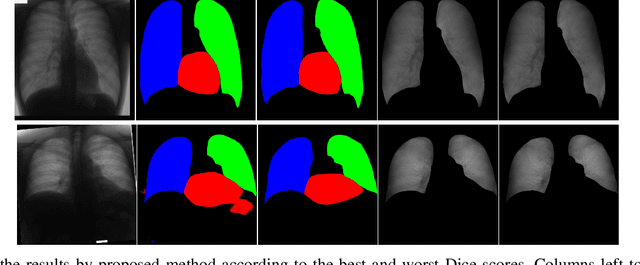
Abstract:Chest X-ray radiography is one of the earliest medical imaging technologies and remains one of the most widely-used for the diagnosis, screening and treatment follow up of diseases related to lungs and heart. The literature in this field of research reports many interesting studies dealing with the challenging tasks of bone suppression and organ segmentation but performed separately, limiting any learning that comes with the consolidation of parameters that could optimize both processes. Although image processing could facilitate computer aided diagnosis, machine learning seems more amenable in dealing with the many parameters one would have to contend with to yield an near optimal classification or decision-making process. This study, and for the first time, introduces a multitask deep learning model that generates simultaneously the bone-suppressed image and the organ segmented image, minimizing as a consequence the number of parameters the model has to deal with and optimizing the processing time as well; while at the same time exploiting the interplay in these parameters so as to benefit the performance of both tasks. The design architecture of this model, which relies on a conditional generative adversarial network, reveals the process on how we managed to modify the well-established pix2pix network to fit the need for multitasking and hence extending the standard image-to-image network to the new image-to-images architecture. Dilated convolutions are also used to improve the results through a more effective receptive field assessment. A comparison of the proposed approach to state-of-the-art algorithms is provided to gauge the merits of the proposed approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge