Farzin Soleymani
CliquePH: Higher-Order Information for Graph Neural Networks through Persistent Homology on Clique Graphs
Sep 12, 2024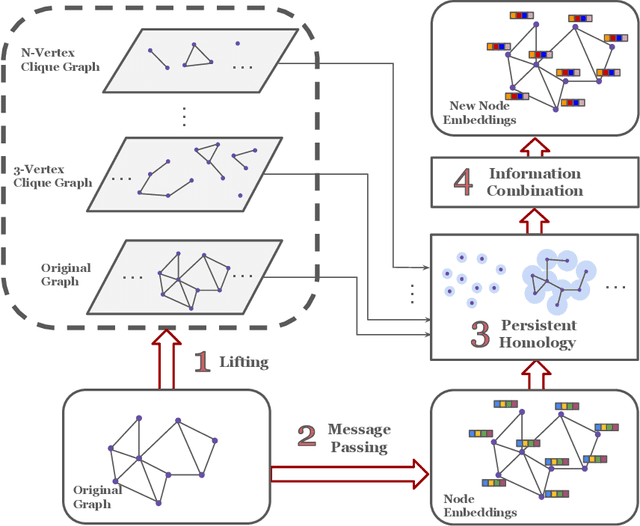
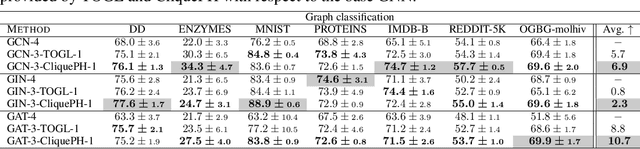
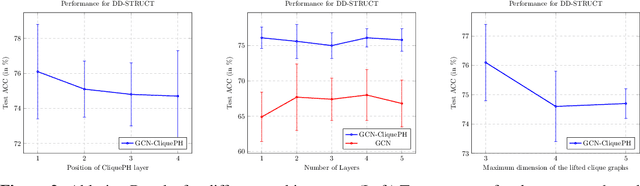

Abstract:Graph neural networks have become the default choice by practitioners for graph learning tasks such as graph classification and node classification. Nevertheless, popular graph neural network models still struggle to capture higher-order information, i.e., information that goes \emph{beyond} pairwise interactions. Recent work has shown that persistent homology, a tool from topological data analysis, can enrich graph neural networks with topological information that they otherwise could not capture. Calculating such features is efficient for dimension 0 (connected components) and dimension 1 (cycles). However, when it comes to higher-order structures, it does not scale well, with a complexity of $O(n^d)$, where $n$ is the number of nodes and $d$ is the order of the structures. In this work, we introduce a novel method that extracts information about higher-order structures in the graph while still using the efficient low-dimensional persistent homology algorithm. On standard benchmark datasets, we show that our method can lead to up to $31\%$ improvements in test accuracy.
Deep Variational Clustering Framework for Self-labeling of Large-scale Medical Images
Sep 22, 2021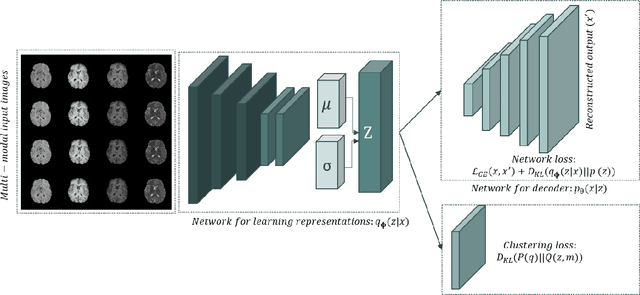
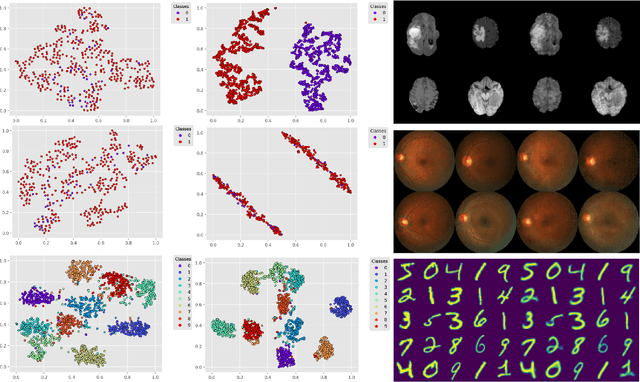
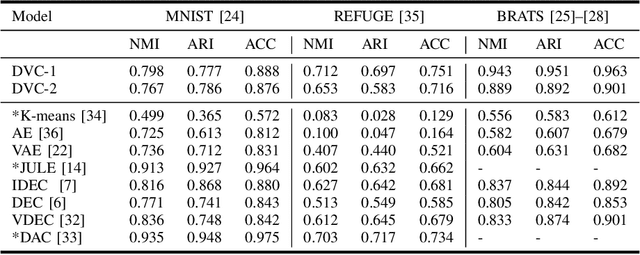
Abstract:We propose a Deep Variational Clustering (DVC) framework for unsupervised representation learning and clustering of large-scale medical images. DVC simultaneously learns the multivariate Gaussian posterior through the probabilistic convolutional encoder and the likelihood distribution with the probabilistic convolutional decoder; and optimizes cluster labels assignment. Here, the learned multivariate Gaussian posterior captures the latent distribution of a large set of unlabeled images. Then, we perform unsupervised clustering on top of the variational latent space using a clustering loss. In this approach, the probabilistic decoder helps to prevent the distortion of data points in the latent space and to preserve the local structure of data generating distribution. The training process can be considered as a self-training process to refine the latent space and simultaneously optimizing cluster assignments iteratively. We evaluated our proposed framework on three public datasets that represented different medical imaging modalities. Our experimental results show that our proposed framework generalizes better across different datasets. It achieves compelling results on several medical imaging benchmarks. Thus, our approach offers potential advantages over conventional deep unsupervised learning in real-world applications. The source code of the method and all the experiments are available publicly at: https://github.com/csfarzin/DVC
Deep Bregman Divergence for Contrastive Learning of Visual Representations
Sep 15, 2021



Abstract:Deep Bregman divergence measures divergence of data points using neural networks which is beyond Euclidean distance and capable of capturing divergence over distributions. In this paper, we propose deep Bregman divergences for contrastive learning of visual representation and we aim to enhance contrastive loss used in self-supervised learning by training additional networks based on functional Bregman divergence. In contrast to the conventional contrastive learning methods which are solely based on divergences between single points, our framework can capture the divergence between distributions which improves the quality of learned representation. By combining conventional contrastive loss with the proposed divergence loss, our method outperforms baseline and most of previous methods for self-supervised and semi-supervised learning on multiple classifications and object detection tasks and datasets. The source code of the method and of all the experiments are available at supplementary.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge