Md Zakir Hossain
SRLoRA: Subspace Recomposition in Low-Rank Adaptation via Importance-Based Fusion and Reinitialization
May 18, 2025Abstract:Low-Rank Adaptation (LoRA) is a widely adopted parameter-efficient fine-tuning (PEFT) method that injects two trainable low-rank matrices (A and B) into frozen pretrained models. While efficient, LoRA constrains updates to a fixed low-rank subspace (Delta W = BA), which can limit representational capacity and hinder downstream performance. We introduce Subspace Recomposition in Low-Rank Adaptation (SRLoRA) via importance-based fusion and reinitialization, a novel approach that enhances LoRA's expressiveness without compromising its lightweight structure. SRLoRA assigns importance scores to each LoRA pair (a column of B and the corresponding row of A), and dynamically recomposes the subspace during training. Less important pairs are fused into the frozen backbone, freeing capacity to reinitialize new pairs along unused principal directions derived from the pretrained weight's singular value decomposition. This mechanism enables continual subspace refreshment and richer adaptation over time, without increasing the number of trainable parameters. We evaluate SRLoRA on both language and vision tasks, including the GLUE benchmark and various image classification datasets. SRLoRA consistently achieves faster convergence and improved accuracy over standard LoRA, demonstrating its generality, efficiency, and potential for broader PEFT applications.
FreqSelect: Frequency-Aware fMRI-to-Image Reconstruction
May 18, 2025Abstract:Reconstructing natural images from functional magnetic resonance imaging (fMRI) data remains a core challenge in natural decoding due to the mismatch between the richness of visual stimuli and the noisy, low resolution nature of fMRI signals. While recent two-stage models, combining deep variational autoencoders (VAEs) with diffusion models, have advanced this task, they treat all spatial-frequency components of the input equally. This uniform treatment forces the model to extract meaning features and suppress irrelevant noise simultaneously, limiting its effectiveness. We introduce FreqSelect, a lightweight, adaptive module that selectively filters spatial-frequency bands before encoding. By dynamically emphasizing frequencies that are most predictive of brain activity and suppressing those that are uninformative, FreqSelect acts as a content-aware gate between image features and natural data. It integrates seamlessly into standard very deep VAE-diffusion pipelines and requires no additional supervision. Evaluated on the Natural Scenes dataset, FreqSelect consistently improves reconstruction quality across both low- and high-level metrics. Beyond performance gains, the learned frequency-selection patterns offer interpretable insights into how different visual frequencies are represented in the brain. Our method generalizes across subjects and scenes, and holds promise for extension to other neuroimaging modalities, offering a principled approach to enhancing both decoding accuracy and neuroscientific interpretability.
Tabular foundation model to detect empathy from visual cues
Apr 15, 2025Abstract:Detecting empathy from video interactions is an emerging area of research. Video datasets, however, are often released as extracted features (i.e., tabular data) rather than raw footage due to privacy and ethical concerns. Prior research on such tabular datasets established tree-based classical machine learning approaches as the best-performing models. Motivated by the recent success of textual foundation models (i.e., large language models), we explore the use of tabular foundation models in empathy detection from tabular visual features. We experiment with two recent tabular foundation models $-$ TabPFN v2 and TabICL $-$ through in-context learning and fine-tuning setups. Our experiments on a public human-robot interaction benchmark demonstrate a significant boost in cross-subject empathy detection accuracy over several strong baselines (accuracy: $0.590 \rightarrow 0.730$; AUC: $0.564 \rightarrow 0.669$). In addition to performance improvement, we contribute novel insights and an evaluation setup to ensure generalisation on unseen subjects in this public benchmark. As the practice of releasing video features as tabular datasets is likely to persist due to privacy constraints, our findings will be widely applicable to future empathy detection video datasets as well.
Unsupervised Search for Ethnic Minorities' Medical Segmentation Training Set
Jan 05, 2025



Abstract:This article investigates the critical issue of dataset bias in medical imaging, with a particular emphasis on racial disparities caused by uneven population distribution in dataset collection. Our analysis reveals that medical segmentation datasets are significantly biased, primarily influenced by the demographic composition of their collection sites. For instance, Scanning Laser Ophthalmoscopy (SLO) fundus datasets collected in the United States predominantly feature images of White individuals, with minority racial groups underrepresented. This imbalance can result in biased model performance and inequitable clinical outcomes, particularly for minority populations. To address this challenge, we propose a novel training set search strategy aimed at reducing these biases by focusing on underrepresented racial groups. Our approach utilizes existing datasets and employs a simple greedy algorithm to identify source images that closely match the target domain distribution. By selecting training data that aligns more closely with the characteristics of minority populations, our strategy improves the accuracy of medical segmentation models on specific minorities, i.e., Black. Our experimental results demonstrate the effectiveness of this approach in mitigating bias. We also discuss the broader societal implications, highlighting how addressing these disparities can contribute to more equitable healthcare outcomes.
Labels Generated by Large Language Model Helps Measuring People's Empathy in Vitro
Jan 01, 2025Abstract:Large language models (LLMs) have revolutionised numerous fields, with LLM-as-a-service (LLMSaaS) having a strong generalisation ability that offers accessible solutions directly without the need for costly training. In contrast to the widely studied prompt engineering for task solving directly (in vivo), this paper explores its potential in in-vitro applications. These involve using LLM to generate labels to help the supervised training of mainstream models by (1) noisy label correction and (2) training data augmentation with LLM-generated labels. In this paper, we evaluate this approach in the emerging field of empathy computing -- automating the prediction of psychological questionnaire outcomes from inputs like text sequences. Specifically, crowdsourced datasets in this domain often suffer from noisy labels that misrepresent underlying empathy. By leveraging LLM-generated labels to train pre-trained language models (PLMs) like RoBERTa, we achieve statistically significant accuracy improvements over baselines, achieving a state-of-the-art Pearson correlation coefficient of 0.648 on NewsEmp benchmarks. In addition, we bring insightful discussions, including current challenges in empathy computing, data biases in training data and evaluation metric selection. Code and LLM-generated data are available at https://github.com/hasan-rakibul/LLMPathy (available once the paper is accepted).
MADE-for-ASD: A Multi-Atlas Deep Ensemble Network for Diagnosing Autism Spectrum Disorder
Jul 09, 2024



Abstract:In response to the global need for efficient early diagnosis of Autism Spectrum Disorder (ASD), this paper bridges the gap between traditional, time-consuming diagnostic methods and potential automated solutions. We propose a multi-atlas deep ensemble network, MADE-for-ASD, that integrates multiple atlases of the brain's functional magnetic resonance imaging (fMRI) data through a weighted deep ensemble network. Our approach integrates demographic information into the prediction workflow, which enhances ASD diagnosis performance and offers a more holistic perspective on patient profiling. We experiment with the well-known publicly available ABIDE (Autism Brain Imaging Data Exchange) I dataset, consisting of resting state fMRI data from 17 different laboratories around the globe. Our proposed system achieves 75.20% accuracy on the entire dataset and 96.40% on a specific subset $-$ both surpassing reported ASD diagnosis accuracy in ABIDE I fMRI studies. Specifically, our model improves by 4.4 percentage points over prior works on the same amount of data. The model exhibits a sensitivity of 82.90% and a specificity of 69.70% on the entire dataset, and 91.00% and 99.50%, respectively, on the specific subset. We leverage the F-score to pinpoint the top 10 ROI in ASD diagnosis, such as \emph{precuneus} and anterior \emph{cingulate/ventromedial}. The proposed system can potentially pave the way for more cost-effective, efficient and scalable strategies in ASD diagnosis. Codes and evaluations are publicly available at TBA.
FunnelNet: An End-to-End Deep Learning Framework to Monitor Digital Heart Murmur in Real-Time
May 10, 2024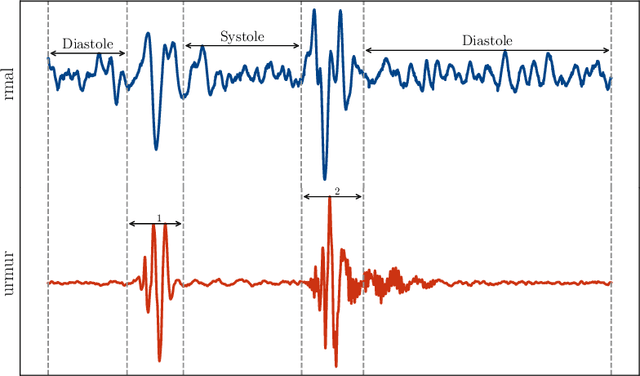
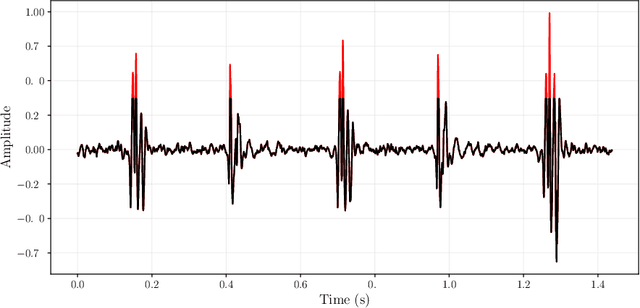
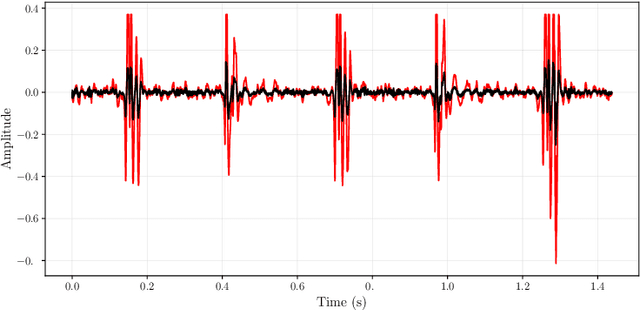
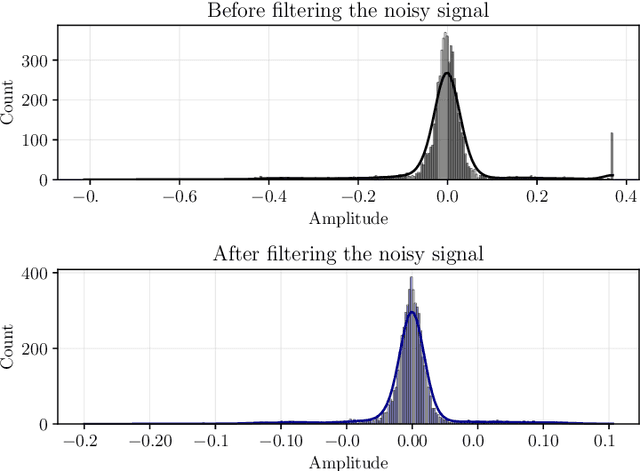
Abstract:Objective: Heart murmurs are abnormal sounds caused by turbulent blood flow within the heart. Several diagnostic methods are available to detect heart murmurs and their severity, such as cardiac auscultation, echocardiography, phonocardiogram (PCG), etc. However, these methods have limitations, including extensive training and experience among healthcare providers, cost and accessibility of echocardiography, as well as noise interference and PCG data processing. This study aims to develop a novel end-to-end real-time heart murmur detection approach using traditional and depthwise separable convolutional networks. Methods: Continuous wavelet transform (CWT) was applied to extract meaningful features from the PCG data. The proposed network has three parts: the Squeeze net, the Bottleneck, and the Expansion net. The Squeeze net generates a compressed data representation, whereas the Bottleneck layer reduces computational complexity using a depthwise-separable convolutional network. The Expansion net is responsible for up-sampling the compressed data to a higher dimension, capturing tiny details of the representative data. Results: For evaluation, we used four publicly available datasets and achieved state-of-the-art performance in all datasets. Furthermore, we tested our proposed network on two resource-constrained devices: a Raspberry PI and an Android device, stripping it down into a tiny machine learning model (TinyML), achieving a maximum of 99.70%. Conclusion: The proposed model offers a deep learning framework for real-time accurate heart murmur detection within limited resources. Significance: It will significantly result in more accessible and practical medical services and reduced diagnosis time to assist medical professionals. The code is publicly available at TBA.
A Deep Learning Approach to Diabetes Diagnosis
Mar 12, 2024



Abstract:Diabetes, resulting from inadequate insulin production or utilization, causes extensive harm to the body. Existing diagnostic methods are often invasive and come with drawbacks, such as cost constraints. Although there are machine learning models like Classwise k Nearest Neighbor (CkNN) and General Regression Neural Network (GRNN), they struggle with imbalanced data and result in under-performance. Leveraging advancements in sensor technology and machine learning, we propose a non-invasive diabetes diagnosis using a Back Propagation Neural Network (BPNN) with batch normalization, incorporating data re-sampling and normalization for class balancing. Our method addresses existing challenges such as limited performance associated with traditional machine learning. Experimental results on three datasets show significant improvements in overall accuracy, sensitivity, and specificity compared to traditional methods. Notably, we achieve accuracies of 89.81% in Pima diabetes dataset, 75.49% in CDC BRFSS2015 dataset, and 95.28% in Mesra Diabetes dataset. This underscores the potential of deep learning models for robust diabetes diagnosis. See project website https://steve-zeyu-zhang.github.io/DiabetesDiagnosis/
Spatial Transcriptomics Analysis of Zero-shot Gene Expression Prediction
Jan 26, 2024



Abstract:Spatial transcriptomics (ST) captures gene expression within distinct regions (i.e., windows) of a tissue slide. Traditional supervised learning frameworks applied to model ST are constrained to predicting expression from slide image windows for gene types seen during training, failing to generalize to unseen gene types. To overcome this limitation, we propose a semantic guided network (SGN), a pioneering zero-shot framework for predicting gene expression from slide image windows. Considering a gene type can be described by functionality and phenotype, we dynamically embed a gene type to a vector per its functionality and phenotype, and employ this vector to project slide image windows to gene expression in feature space, unleashing zero-shot expression prediction for unseen gene types. The gene type functionality and phenotype are queried with a carefully designed prompt from a pre-trained large language model (LLM). On standard benchmark datasets, we demonstrate competitive zero-shot performance compared to past state-of-the-art supervised learning approaches.
Empathy Detection Using Machine Learning on Text, Audiovisual, Audio or Physiological Signals
Oct 30, 2023Abstract:Empathy is a social skill that indicates an individual's ability to understand others. Over the past few years, empathy has drawn attention from various disciplines, including but not limited to Affective Computing, Cognitive Science and Psychology. Empathy is a context-dependent term; thus, detecting or recognising empathy has potential applications in society, healthcare and education. Despite being a broad and overlapping topic, the avenue of empathy detection studies leveraging Machine Learning remains underexplored from a holistic literature perspective. To this end, we systematically collect and screen 801 papers from 10 well-known databases and analyse the selected 54 papers. We group the papers based on input modalities of empathy detection systems, i.e., text, audiovisual, audio and physiological signals. We examine modality-specific pre-processing and network architecture design protocols, popular dataset descriptions and availability details, and evaluation protocols. We further discuss the potential applications, deployment challenges and research gaps in the Affective Computing-based empathy domain, which can facilitate new avenues of exploration. We believe that our work is a stepping stone to developing a privacy-preserving and unbiased empathic system inclusive of culture, diversity and multilingualism that can be deployed in practice to enhance the overall well-being of human life.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge