Manfred Huber
Reducing Label Dependency in Human Activity Recognition with Wearables: From Supervised Learning to Novel Weakly Self-Supervised Approaches
Dec 15, 2025Abstract:Human activity recognition (HAR) using wearable sensors has advanced through various machine learning paradigms, each with inherent trade-offs between performance and labeling requirements. While fully supervised techniques achieve high accuracy, they demand extensive labeled datasets that are costly to obtain. Conversely, unsupervised methods eliminate labeling needs but often deliver suboptimal performance. This paper presents a comprehensive investigation across the supervision spectrum for wearable-based HAR, with particular focus on novel approaches that minimize labeling requirements while maintaining competitive accuracy. We develop and empirically compare: (1) traditional fully supervised learning, (2) basic unsupervised learning, (3) a weakly supervised learning approach with constraints, (4) a multi-task learning approach with knowledge sharing, (5) a self-supervised approach based on domain expertise, and (6) a novel weakly self-supervised learning framework that leverages domain knowledge and minimal labeled data. Experiments across benchmark datasets demonstrate that: (i) our weakly supervised methods achieve performance comparable to fully supervised approaches while significantly reducing supervision requirements; (ii) the proposed multi-task framework enhances performance through knowledge sharing between related tasks; (iii) our weakly self-supervised approach demonstrates remarkable efficiency with just 10\% of labeled data. These results not only highlight the complementary strengths of different learning paradigms, offering insights into tailoring HAR solutions based on the availability of labeled data, but also establish that our novel weakly self-supervised framework offers a promising solution for practical HAR applications where labeled data are limited.
DeGuV: Depth-Guided Visual Reinforcement Learning for Generalization and Interpretability in Manipulation
Sep 05, 2025Abstract:Reinforcement learning (RL) agents can learn to solve complex tasks from visual inputs, but generalizing these learned skills to new environments remains a major challenge in RL application, especially robotics. While data augmentation can improve generalization, it often compromises sample efficiency and training stability. This paper introduces DeGuV, an RL framework that enhances both generalization and sample efficiency. In specific, we leverage a learnable masker network that produces a mask from the depth input, preserving only critical visual information while discarding irrelevant pixels. Through this, we ensure that our RL agents focus on essential features, improving robustness under data augmentation. In addition, we incorporate contrastive learning and stabilize Q-value estimation under augmentation to further enhance sample efficiency and training stability. We evaluate our proposed method on the RL-ViGen benchmark using the Franka Emika robot and demonstrate its effectiveness in zero-shot sim-to-real transfer. Our results show that DeGuV outperforms state-of-the-art methods in both generalization and sample efficiency while also improving interpretability by highlighting the most relevant regions in the visual input
FlowMP: Learning Motion Fields for Robot Planning with Conditional Flow Matching
Mar 08, 2025Abstract:Prior flow matching methods in robotics have primarily learned velocity fields to morph one distribution of trajectories into another. In this work, we extend flow matching to capture second-order trajectory dynamics, incorporating acceleration effects either explicitly in the model or implicitly through the learning objective. Unlike diffusion models, which rely on a noisy forward process and iterative denoising steps, flow matching trains a continuous transformation (flow) that directly maps a simple prior distribution to the target trajectory distribution without any denoising procedure. By modeling trajectories with second-order dynamics, our approach ensures that generated robot motions are smooth and physically executable, avoiding the jerky or dynamically infeasible trajectories that first-order models might produce. We empirically demonstrate that this second-order conditional flow matching yields superior performance on motion planning benchmarks, achieving smoother trajectories and higher success rates than baseline planners. These findings highlight the advantage of learning acceleration-aware motion fields, as our method outperforms existing motion planning methods in terms of trajectory quality and planning success.
Volumetric Mapping with Panoptic Refinement via Kernel Density Estimation for Mobile Robots
Dec 15, 2024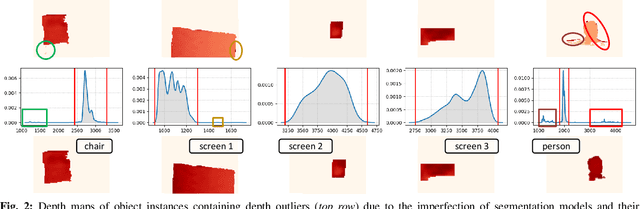
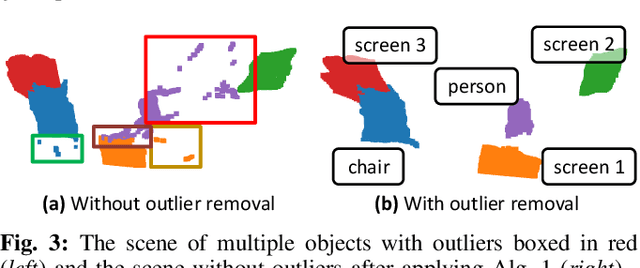


Abstract:Reconstructing three-dimensional (3D) scenes with semantic understanding is vital in many robotic applications. Robots need to identify which objects, along with their positions and shapes, to manipulate them precisely with given tasks. Mobile robots, especially, usually use lightweight networks to segment objects on RGB images and then localize them via depth maps; however, they often encounter out-of-distribution scenarios where masks over-cover the objects. In this paper, we address the problem of panoptic segmentation quality in 3D scene reconstruction by refining segmentation errors using non-parametric statistical methods. To enhance mask precision, we map the predicted masks into a depth frame to estimate their distribution via kernel densities. The outliers in depth perception are then rejected without the need for additional parameters in an adaptive manner to out-of-distribution scenarios, followed by 3D reconstruction using projective signed distance functions (SDFs). We validate our method on a synthetic dataset, which shows improvements in both quantitative and qualitative results for panoptic mapping. Through real-world testing, the results furthermore show our method's capability to be deployed on a real-robot system. Our source code is available at: https://github.com/mkhangg/refined panoptic mapping.
Weakly Supervised Multi-Task Representation Learning for Human Activity Analysis Using Wearables
Aug 06, 2023Abstract:Sensor data streams from wearable devices and smart environments are widely studied in areas like human activity recognition (HAR), person identification, or health monitoring. However, most of the previous works in activity and sensor stream analysis have been focusing on one aspect of the data, e.g. only recognizing the type of the activity or only identifying the person who performed the activity. We instead propose an approach that uses a weakly supervised multi-output siamese network that learns to map the data into multiple representation spaces, where each representation space focuses on one aspect of the data. The representation vectors of the data samples are positioned in the space such that the data with the same semantic meaning in that aspect are closely located to each other. Therefore, as demonstrated with a set of experiments, the trained model can provide metrics for clustering data based on multiple aspects, allowing it to address multiple tasks simultaneously and even to outperform single task supervised methods in many situations. In addition, further experiments are presented that in more detail analyze the effect of the architecture and of using multiple tasks within this framework, that investigate the scalability of the model to include additional tasks, and that demonstrate the ability of the framework to combine data for which only partial relationship information with respect to the target tasks is available.
Unsupervised Embedding Learning for Human Activity Recognition Using Wearable Sensor Data
Jul 21, 2023Abstract:The embedded sensors in widely used smartphones and other wearable devices make the data of human activities more accessible. However, recognizing different human activities from the wearable sensor data remains a challenging research problem in ubiquitous computing. One of the reasons is that the majority of the acquired data has no labels. In this paper, we present an unsupervised approach, which is based on the nature of human activity, to project the human activities into an embedding space in which similar activities will be located closely together. Using this, subsequent clustering algorithms can benefit from the embeddings, forming behavior clusters that represent the distinct activities performed by a person. Results of experiments on three labeled benchmark datasets demonstrate the effectiveness of the framework and show that our approach can help the clustering algorithm achieve improved performance in identifying and categorizing the underlying human activities compared to unsupervised techniques applied directly to the original data set.
Siamese Networks for Weakly Supervised Human Activity Recognition
Jul 18, 2023Abstract:Deep learning has been successfully applied to human activity recognition. However, training deep neural networks requires explicitly labeled data which is difficult to acquire. In this paper, we present a model with multiple siamese networks that are trained by using only the information about the similarity between pairs of data samples without knowing the explicit labels. The trained model maps the activity data samples into fixed size representation vectors such that the distance between the vectors in the representation space approximates the similarity of the data samples in the input space. Thus, the trained model can work as a metric for a wide range of different clustering algorithms. The training process minimizes a similarity loss function that forces the distance metric to be small for pairs of samples from the same kind of activity, and large for pairs of samples from different kinds of activities. We evaluate the model on three datasets to verify its effectiveness in segmentation and recognition of continuous human activity sequences.
Histopathology Slide Indexing and Search: Are We There Yet?
Jun 29, 2023
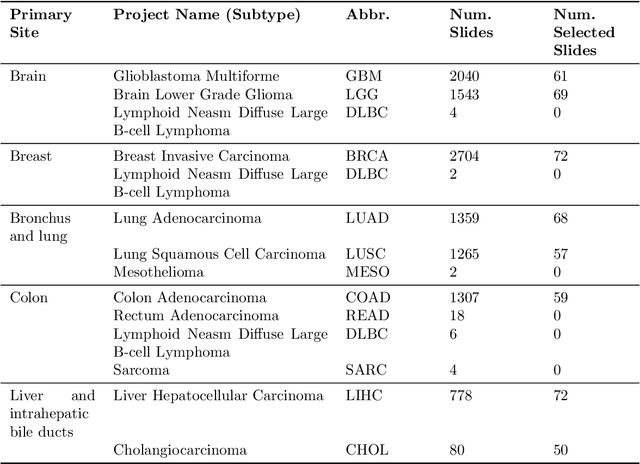


Abstract:The search and retrieval of digital histopathology slides is an important task that has yet to be solved. In this case study, we investigate the clinical readiness of three state-of-the-art histopathology slide search engines, Yottixel, SISH, and RetCCL, on three patients with solid tumors. We provide a qualitative assessment of each model's performance in providing retrieval results that are reliable and useful to pathologists. We found that all three image search engines fail to produce consistently reliable results and have difficulties in capturing granular and subtle features of malignancy, limiting their diagnostic accuracy. Based on our findings, we also propose a minimal set of requirements to further advance the development of accurate and reliable histopathology image search engines for successful clinical adoption.
ExtPerFC: An Efficient 2D and 3D Perception Hardware-Software Framework for Mobile Cobot
Jun 08, 2023Abstract:As the reliability of the robot's perception correlates with the number of integrated sensing modalities to tackle uncertainty, a practical solution to manage these sensors from different computers, operate them simultaneously, and maintain their real-time performance on the existing robotic system with minimal effort is needed. In this work, we present an end-to-end software-hardware framework, namely ExtPerFC, that supports both conventional hardware and software components and integrates machine learning object detectors without requiring an additional dedicated graphic processor unit (GPU). We first design our framework to achieve real-time performance on the existing robotic system, guarantee configuration optimization, and concentrate on code reusability. We then mathematically model and utilize our transfer learning strategies for 2D object detection and fuse them into depth images for 3D depth estimation. Lastly, we systematically test the proposed framework on the Baxter robot with two 7-DOF arms, a four-wheel mobility base, and an Intel RealSense D435i RGB-D camera. The results show that the robot achieves real-time performance while executing other tasks (e.g., map building, localization, navigation, object detection, arm moving, and grasping) simultaneously with available hardware like Intel onboard CPUS/GPUs on distributed computers. Also, to comprehensively control, program, and monitor the robot system, we design and introduce an end-user application. The source code is available at https://github.com/tuantdang/perception_framework.
Clinically Relevant Latent Space Embedding of Cancer Histopathology Slides through Variational Autoencoder Based Image Compression
Mar 23, 2023Abstract:In this paper, we introduce a Variational Autoencoder (VAE) based training approach that can compress and decompress cancer pathology slides at a compression ratio of 1:512, which is better than the previously reported state of the art (SOTA) in the literature, while still maintaining accuracy in clinical validation tasks. The compression approach was tested on more common computer vision datasets such as CIFAR10, and we explore which image characteristics enable this compression ratio on cancer imaging data but not generic images. We generate and visualize embeddings from the compressed latent space and demonstrate how they are useful for clinical interpretation of data, and how in the future such latent embeddings can be used to accelerate search of clinical imaging data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge