Parisa Boodaghi Malidarreh
Dual Codebook VQ: Enhanced Image Reconstruction with Reduced Codebook Size
Mar 13, 2025Abstract:Vector Quantization (VQ) techniques face significant challenges in codebook utilization, limiting reconstruction fidelity in image modeling. We introduce a Dual Codebook mechanism that effectively addresses this limitation by partitioning the representation into complementary global and local components. The global codebook employs a lightweight transformer for concurrent updates of all code vectors, while the local codebook maintains precise feature representation through deterministic selection. This complementary approach is trained from scratch without requiring pre-trained knowledge. Experimental evaluation across multiple standard benchmark datasets demonstrates state-of-the-art reconstruction quality while using a compact codebook of size 512 - half the size of previous methods that require pre-training. Our approach achieves significant FID improvements across diverse image domains, particularly excelling in scene and face reconstruction tasks. These results establish Dual Codebook VQ as an efficient paradigm for high-fidelity image reconstruction with significantly reduced computational requirements.
Predicting Future States with Spatial Point Processes in Single Molecule Resolution Spatial Transcriptomics
Jan 04, 2024



Abstract:In this paper, we introduce a pipeline based on Random Forest Regression to predict the future distribution of cells that are expressed by the Sog-D gene (active cells) in both the Anterior to posterior (AP) and the Dorsal to Ventral (DV) axis of the Drosophila in embryogenesis process. This method provides insights about how cells and living organisms control gene expression in super resolution whole embryo spatial transcriptomics imaging at sub cellular, single molecule resolution. A Random Forest Regression model was used to predict the next stage active distribution based on the previous one. To achieve this goal, we leveraged temporally resolved, spatial point processes by including Ripley's K-function in conjunction with the cell's state in each stage of embryogenesis, and found average predictive accuracy of active cell distribution. This tool is analogous to RNA Velocity for spatially resolved developmental biology, from one data point we can predict future spatially resolved gene expression using features from the spatial point processes.
Real-Time Diagnostic Integrity Meets Efficiency: A Novel Platform-Agnostic Architecture for Physiological Signal Compression
Jan 04, 2024Abstract:Head-based signals such as EEG, EMG, EOG, and ECG collected by wearable systems will play a pivotal role in clinical diagnosis, monitoring, and treatment of important brain disorder diseases. However, the real-time transmission of the significant corpus physiological signals over extended periods consumes substantial power and time, limiting the viability of battery-dependent physiological monitoring wearables. This paper presents a novel deep-learning framework employing a variational autoencoder (VAE) for physiological signal compression to reduce wearables' computational complexity and energy consumption. Our approach achieves an impressive compression ratio of 1:293 specifically for spectrogram data, surpassing state-of-the-art compression techniques such as JPEG2000, H.264, Direct Cosine Transform (DCT), and Huffman Encoding, which do not excel in handling physiological signals. We validate the efficacy of the compressed algorithms using collected physiological signals from real patients in the Hospital and deploy the solution on commonly used embedded AI chips (i.e., ARM Cortex V8 and Jetson Nano). The proposed framework achieves a 91% seizure detection accuracy using XGBoost, confirming the approach's reliability, practicality, and scalability.
The State of Applying Artificial Intelligence to Tissue Imaging for Cancer Research and Early Detection
Jun 29, 2023



Abstract:Artificial intelligence represents a new frontier in human medicine that could save more lives and reduce the costs, thereby increasing accessibility. As a consequence, the rate of advancement of AI in cancer medical imaging and more particularly tissue pathology has exploded, opening it to ethical and technical questions that could impede its adoption into existing systems. In order to chart the path of AI in its application to cancer tissue imaging, we review current work and identify how it can improve cancer pathology diagnostics and research. In this review, we identify 5 core tasks that models are developed for, including regression, classification, segmentation, generation, and compression tasks. We address the benefits and challenges that such methods face, and how they can be adapted for use in cancer prevention and treatment. The studies looked at in this paper represent the beginning of this field and future experiments will build on the foundations that we highlight.
Histopathology Slide Indexing and Search: Are We There Yet?
Jun 29, 2023
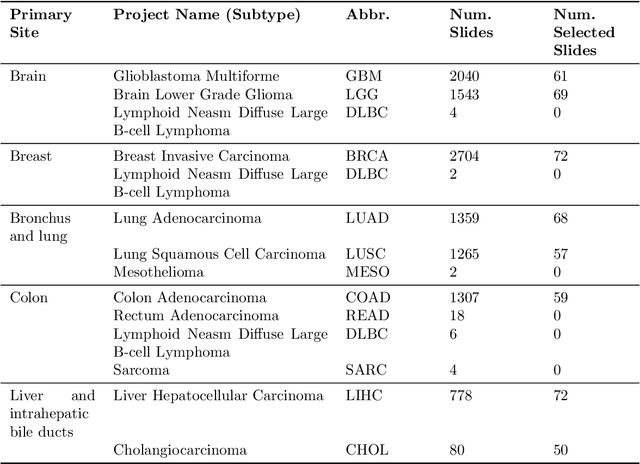


Abstract:The search and retrieval of digital histopathology slides is an important task that has yet to be solved. In this case study, we investigate the clinical readiness of three state-of-the-art histopathology slide search engines, Yottixel, SISH, and RetCCL, on three patients with solid tumors. We provide a qualitative assessment of each model's performance in providing retrieval results that are reliable and useful to pathologists. We found that all three image search engines fail to produce consistently reliable results and have difficulties in capturing granular and subtle features of malignancy, limiting their diagnostic accuracy. Based on our findings, we also propose a minimal set of requirements to further advance the development of accurate and reliable histopathology image search engines for successful clinical adoption.
Multimodal Pathology Image Search Between H&E Slides and Multiplexed Immunofluorescent Images
Jun 11, 2023



Abstract:We present an approach for multimodal pathology image search, using dynamic time warping (DTW) on Variational Autoencoder (VAE) latent space that is fed into a ranked choice voting scheme to retrieve multiplexed immunofluorescent imaging (mIF) that is most similar to a query H&E slide. Through training the VAE and applying DTW, we align and compare mIF and H&E slides. Our method improves differential diagnosis and therapeutic decisions by integrating morphological H&E data with immunophenotyping from mIF, providing clinicians a rich perspective of disease states. This facilitates an understanding of the spatial relationships in tissue samples and could revolutionize the diagnostic process, enhancing precision and enabling personalized therapy selection. Our technique demonstrates feasibility using colorectal cancer and healthy tonsil samples. An exhaustive ablation study was conducted on a search engine designed to explore the correlation between multiplexed Immunofluorescence (mIF) and Hematoxylin and Eosin (H&E) staining, in order to validate its ability to map these distinct modalities into a unified vector space. Despite extreme class imbalance, the system demonstrated robustness and utility by returning similar results across various data features, which suggests potential for future use in multimodal histopathology data analysis.
Clinically Relevant Latent Space Embedding of Cancer Histopathology Slides through Variational Autoencoder Based Image Compression
Mar 23, 2023Abstract:In this paper, we introduce a Variational Autoencoder (VAE) based training approach that can compress and decompress cancer pathology slides at a compression ratio of 1:512, which is better than the previously reported state of the art (SOTA) in the literature, while still maintaining accuracy in clinical validation tasks. The compression approach was tested on more common computer vision datasets such as CIFAR10, and we explore which image characteristics enable this compression ratio on cancer imaging data but not generic images. We generate and visualize embeddings from the compressed latent space and demonstrate how they are useful for clinical interpretation of data, and how in the future such latent embeddings can be used to accelerate search of clinical imaging data.
An Artificial Neural Network-Based Model Predictive Control for Three-phase Flying Capacitor Multi-Level Inverter
Oct 15, 2021



Abstract:Model predictive control (MPC) has been used widely in power electronics due to its simple concept, fast dynamic response, and good reference tracking. However, it suffers from parametric uncertainties, since it directly relies on the mathematical model of the system to predict the optimal switching states to be used at the next sampling time. As a result, uncertain parameters lead to an ill-designed MPC. Thus, this paper offers a model-free control strategy on the basis of artificial neural networks (ANNs), for mitigating the effects of parameter mismatching while having a little negative impact on the inverter's performance. This method includes two related stages. First, MPC is used as an expert to control the studied converter in order to provide the training data; while, in the second stage, the obtained dataset is utilized to train the proposed ANN which will be used directly to control the inverter without the requirement for the mathematical model of the system. The case study herein is based on a four-level three-cell flying capacitor inverter. In this study, MATLAB/Simulink is used to simulate the performance of the proposed control strategy, taking into account various operating conditions. Afterward, the simulation results are reported in comparison with the conventional MPC scheme, demonstrating the superior performance of the proposed control strategy in terms of getting low total harmonic distortion (THD) and the robustness against parameters mismatch, especially when changes occur in the system parameters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge