Luiz E. S. Oliveira
Automatic Chronic Degenerative Diseases Identification Using Enteric Nervous System Images
Oct 31, 2020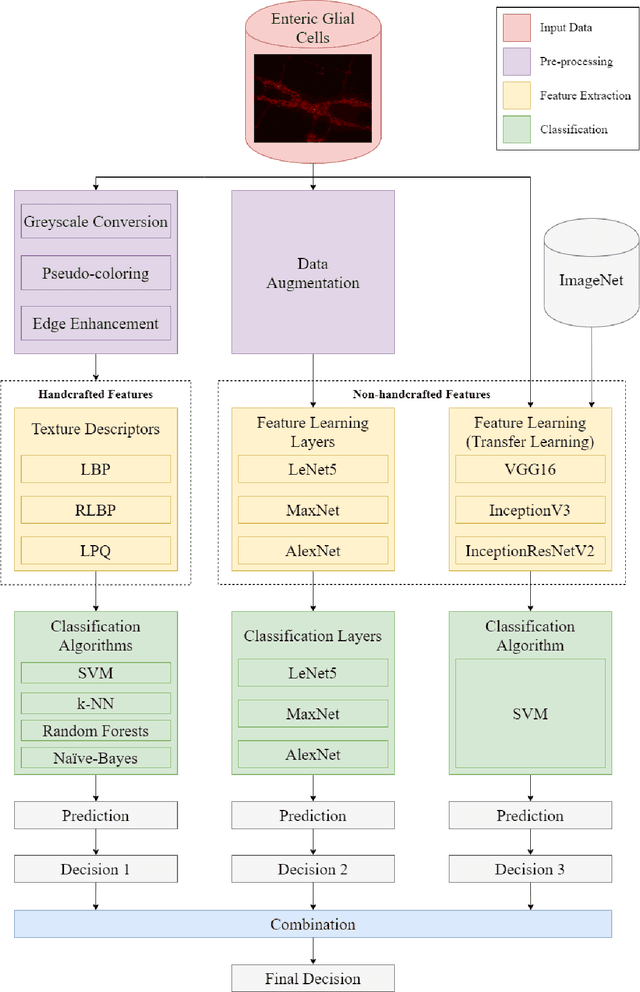

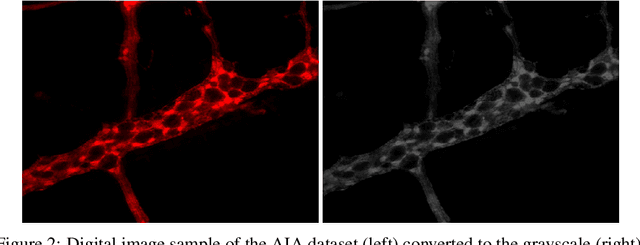
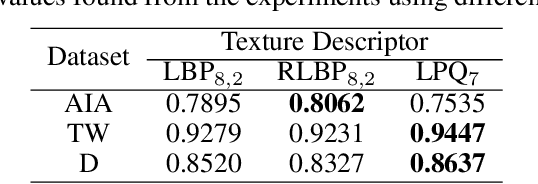
Abstract:Studies recently accomplished on the Enteric Nervous System have shown that chronic degenerative diseases affect the Enteric Glial Cells (EGC) and, thus, the development of recognition methods able to identify whether or not the EGC are affected by these type of diseases may be helpful in its diagnoses. In this work, we propose the use of pattern recognition and machine learning techniques to evaluate if a given animal EGC image was obtained from a healthy individual or one affect by a chronic degenerative disease. In the proposed approach, we have performed the classification task with handcrafted features and deep learning based techniques, also known as non-handcrafted features. The handcrafted features were obtained from the textural content of the ECG images using texture descriptors, such as the Local Binary Pattern (LBP). Moreover, the representation learning techniques employed in the approach are based on different Convolutional Neural Network (CNN) architectures, such as AlexNet and VGG16, with and without transfer learning. The complementarity between the handcrafted and non-handcrafted features was also evaluated with late fusion techniques. The datasets of EGC images used in the experiments, which are also contributions of this paper, are composed of three different chronic degenerative diseases: Cancer, Diabetes Mellitus, and Rheumatoid Arthritis. The experimental results, supported by statistical analysis, shown that the proposed approach can distinguish healthy cells from the sick ones with a recognition rate of 89.30% (Rheumatoid Arthritis), 98.45% (Cancer), and 95.13% (Diabetes Mellitus), being achieved by combining classifiers obtained both feature scenarios.
Two-View Fine-grained Classification of Plant Species
May 18, 2020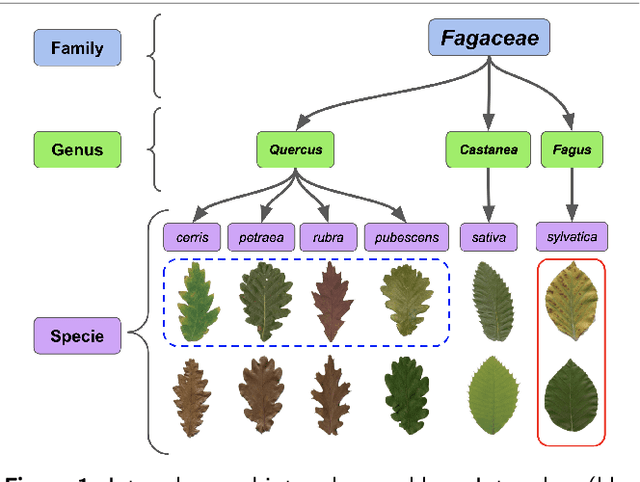

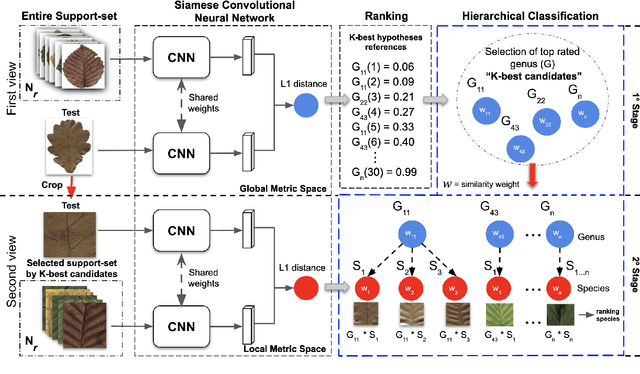

Abstract:Automatic plant classification is a challenging problem due to the wide biodiversity of the existing plant species in a fine-grained scenario. Powerful deep learning architectures have been used to improve the classification performance in such a fine-grained problem, but usually building models that are highly dependent on a large training dataset and which are not scalable. In this paper, we propose a novel method based on a two-view leaf image representation and a hierarchical classification strategy for fine-grained recognition of plant species. It uses the botanical taxonomy as a basis for a coarse-to-fine strategy applied to identify the plant genus and species. The two-view representation provides complementary global and local features of leaf images. A deep metric based on Siamese convolutional neural networks is used to reduce the dependence on a large number of training samples and make the method scalable to new plant species. The experimental results on two challenging fine-grained datasets of leaf images (i.e. LifeCLEF 2015 and LeafSnap) have shown the effectiveness of the proposed method, which achieved recognition accuracy of 0.87 and 0.96 respectively.
An End-to-End Approach for Recognition of Modern and Historical Handwritten Numeral Strings
Mar 28, 2020
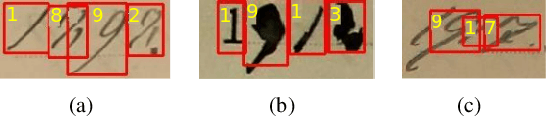

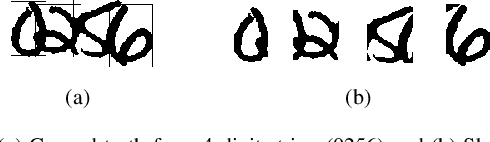
Abstract:An end-to-end solution for handwritten numeral string recognition is proposed, in which the numeral string is considered as composed of objects automatically detected and recognized by a YoLo-based model. The main contribution of this paper is to avoid heuristic-based methods for string preprocessing and segmentation, the need for task-oriented classifiers, and also the use of specific constraints related to the string length. A robust experimental protocol based on several numeral string datasets, including one composed of historical documents, has shown that the proposed method is a feasible end-to-end solution for numeral string recognition. Besides, it reduces the complexity of the string recognition task considerably since it drops out classical steps, in special preprocessing, segmentation, and a set of classifiers devoted to strings with a specific length.
Data Augmentation for Histopathological Images Based on Gaussian-Laplacian Pyramid Blending
Jan 31, 2020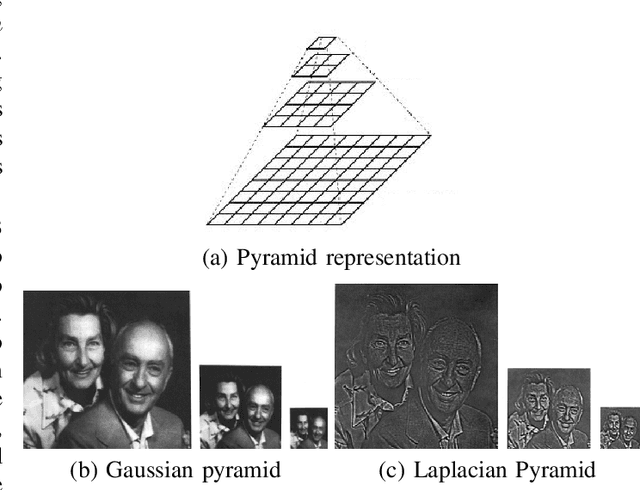
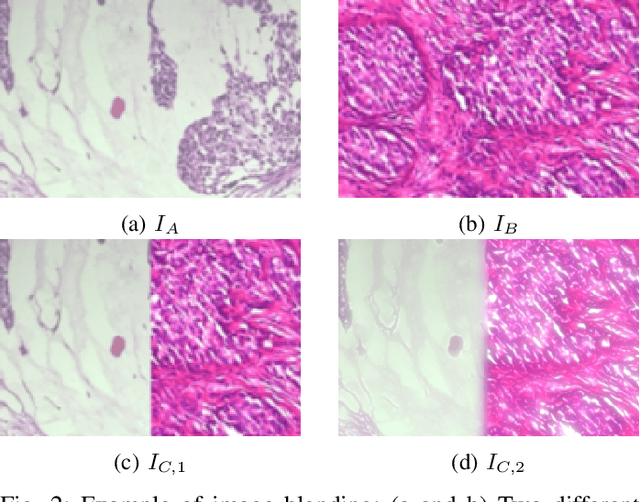
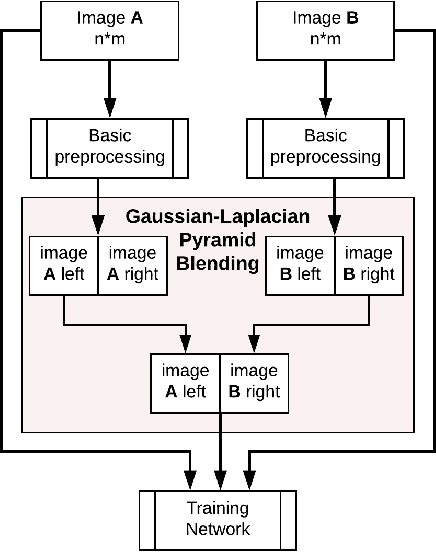
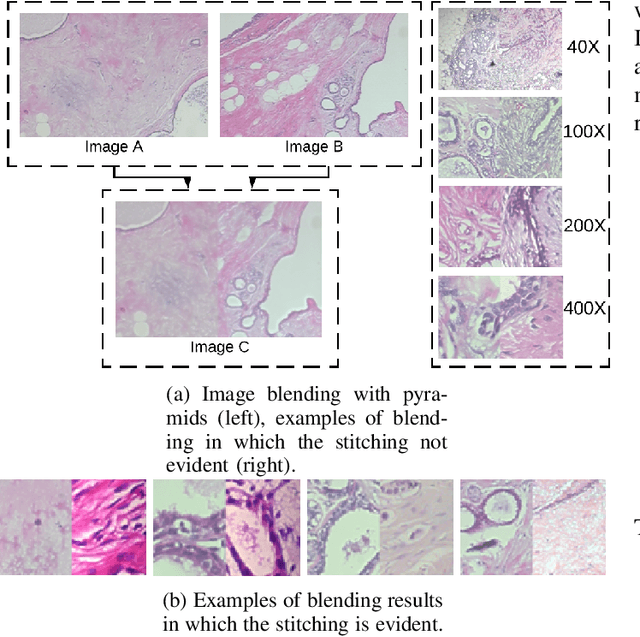
Abstract:Data imbalance is a major problem that affects several machine learning algorithms. Such problems are troublesome because most of the learning algorithms attempts to optimize a loss function based on error measures that do not take into account the data imbalance. Accordingly, the learning algorithm simply generates a trivial model that is biased toward predicting the most frequent class in the training data. Data augmentation techniques have been used to mitigate the data imbalance problem. However, in the case of histopathologic images (HIs), low-level as well as high-level data augmentation techniques still present performance issues when applied in the presence of inter-patient variability; whence the model tends to learn color representations, which are in fact related to the stain process. In this paper, we propose an approach capable of not only augmenting HIs database but also distributing the inter-patient variability by means of image blending using Gaussian-Laplacian pyramid. The proposed approach consists in finding the Gaussian pyramids of two images of different patients and finding the Laplacian pyramids thereof. Afterwards, the left half of one image and the right half of another are joined in each level of Laplacian pyramid, and from the joint pyramids, the original image is reconstructed. This composition, resulting from the blending process, combines stain variation of two patients, avoiding that color misleads the learning process. Experimental results on the BreakHis dataset have shown promising gains vis-\`a-vis the majority of traditional techniques presented in the literature.
Histopathologic Image Processing: A Review
Apr 16, 2019
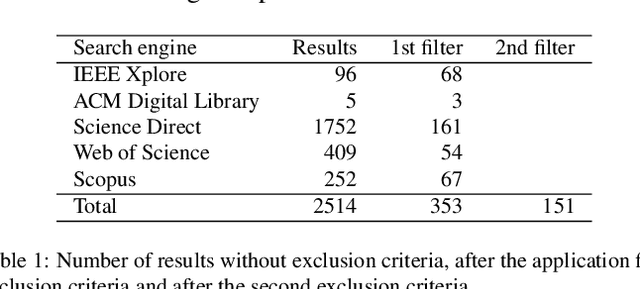

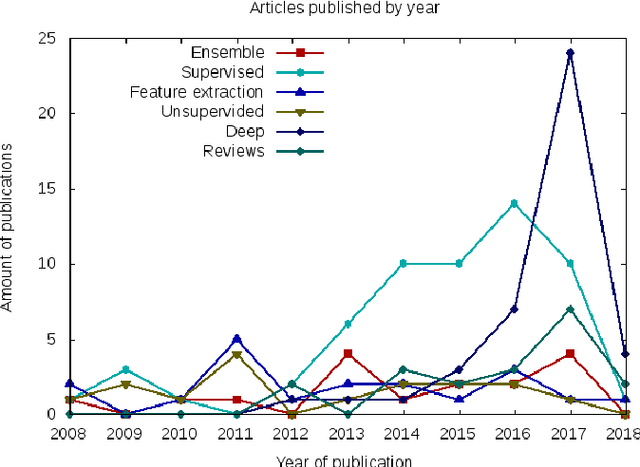
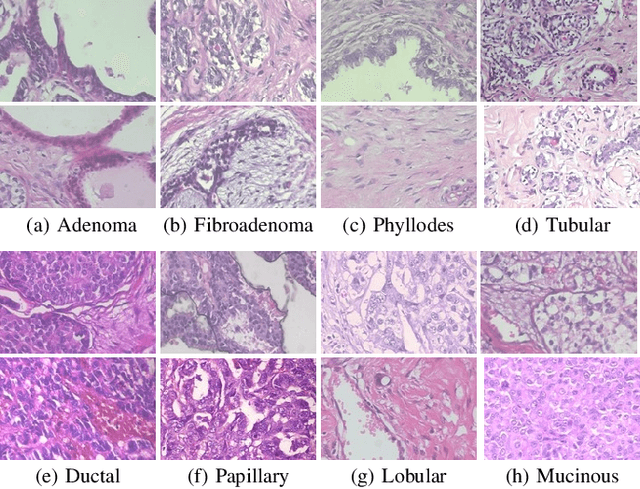
Abstract:Histopathologic Images (HI) are the gold standard for evaluation of some tumors. However, the analysis of such images is challenging even for experienced pathologists, resulting in problems of inter and intra observer. Besides that, the analysis is time and resource consuming. One of the ways to accelerate such an analysis is by using Computer Aided Diagnosis systems. In this work we present a literature review about the computing techniques to process HI, including shallow and deep methods. We cover the most common tasks for processing HI such as segmentation, feature extraction, unsupervised learning and supervised learning. A dataset section show some datasets found during the literature review. We also bring a study case of breast cancer classification using a mix of deep and shallow machine learning methods. The proposed method obtained an accuracy of 91% in the best case, outperforming the compared baseline of the dataset.
Double Transfer Learning for Breast Cancer Histopathologic Image Classification
Apr 16, 2019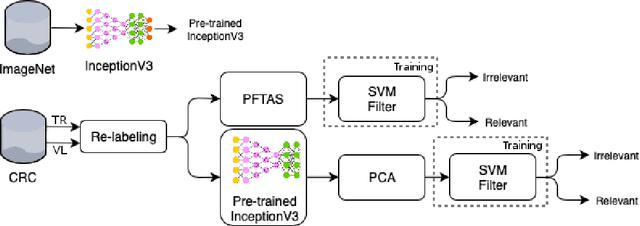
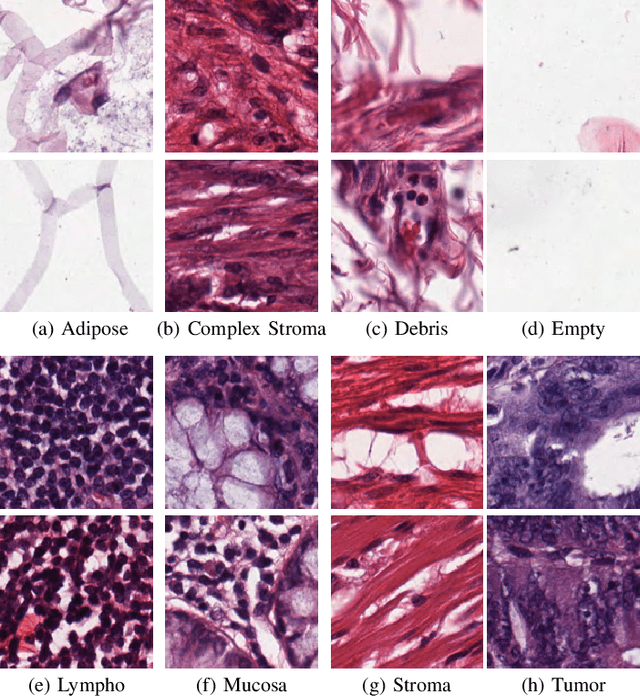
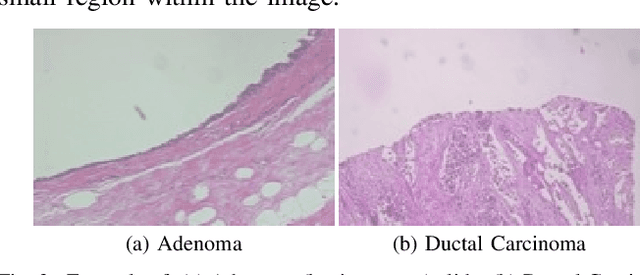
Abstract:This work proposes a classification approach for breast cancer histopathologic images (HI) that uses transfer learning to extract features from HI using an Inception-v3 CNN pre-trained with ImageNet dataset. We also use transfer learning on training a support vector machine (SVM) classifier on a tissue labeled colorectal cancer dataset aiming to filter the patches from a breast cancer HI and remove the irrelevant ones. We show that removing irrelevant patches before training a second SVM classifier, improves the accuracy for classifying malign and benign tumors on breast cancer images. We are able to improve the classification accuracy in 3.7% using the feature extraction transfer learning and an additional 0.7% using the irrelevant patch elimination. The proposed approach outperforms the state-of-the-art in three out of the four magnification factors of the breast cancer dataset.
A Classifier-free Ensemble Selection Method based on Data Diversity in Random Subspaces
Aug 13, 2014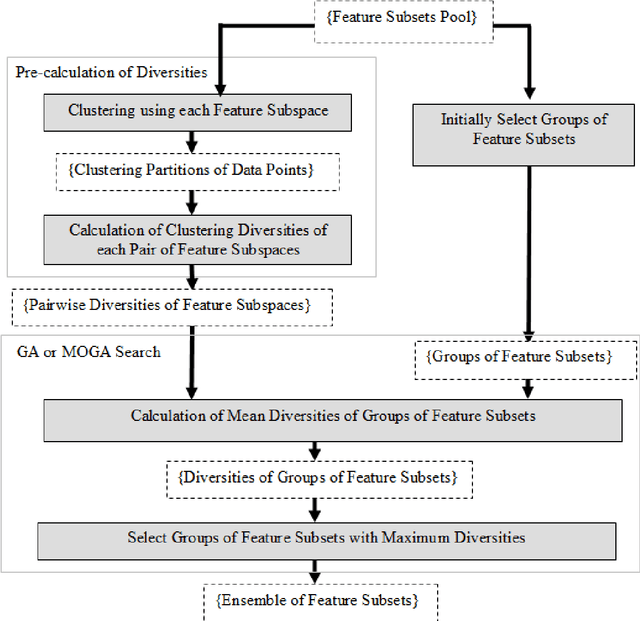
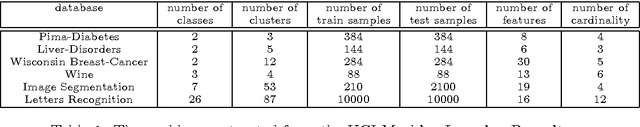

Abstract:The Ensemble of Classifiers (EoC) has been shown to be effective in improving the performance of single classifiers by combining their outputs, and one of the most important properties involved in the selection of the best EoC from a pool of classifiers is considered to be classifier diversity. In general, classifier diversity does not occur randomly, but is generated systematically by various ensemble creation methods. By using diverse data subsets to train classifiers, these methods can create diverse classifiers for the EoC. In this work, we propose a scheme to measure data diversity directly from random subspaces, and explore the possibility of using it to select the best data subsets for the construction of the EoC. Our scheme is the first ensemble selection method to be presented in the literature based on the concept of data diversity. Its main advantage over the traditional framework (ensemble creation then selection) is that it obviates the need for classifier training prior to ensemble selection. A single Genetic Algorithm (GA) and a Multi-Objective Genetic Algorithm (MOGA) were evaluated to search for the best solutions for the classifier-free ensemble selection. In both cases, objective functions based on different clustering diversity measures were implemented and tested. All the results obtained with the proposed classifier-free ensemble selection method were compared with the traditional classifier-based ensemble selection using Mean Classifier Error (ME) and Majority Voting Error (MVE). The applicability of the method is tested on UCI machine learning problems and NIST SD19 handwritten numerals.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge